Damaged proteins and proteins that have finished their role in the cell must be removed and broken down, but at the same time care must be taken that essential proteins are not accidentally sent to "shredding"

Damaged proteins and proteins that have finished their role in the cell must be removed and broken down, but at the same time care must be taken that essential proteins are not accidentally sent to "shredding". A new study by the institute's scientists, led by Dr. Michal Sharon from the Department of Biological Chemistry, reveals how the "rescue" enzyme works, which prevents unwanted breakdown of essential proteins. This enzyme rescues, among other things, proteins that suppress cancer tumors - such as p53 and its lesser-known brother, p73 - and thus protects against cancer.
The breakdown of damaged proteins, or proteins that have finished their function, is done through an orderly and controlled "bureaucratic" process, in which the proteins that are candidates for breakdown are marked with a special label - ubiquitin - and transferred to the "degradation wing", the proteasome. After identifying the label - which is done using a certain unit of the proteasome - the protein enters another unit, which functions as a "shredder", where it is broken down into its components. However, apart from the "bureaucratic" process, the discovery of which won Rose, Hershko and Czechnoover the Nobel Prize in Chemistry for 2004, the importance of another, passive and more "spontaneous" decomposition process has become increasingly clear in recent years. It turns out that certain proteins - which lack a folded and ordered structure, in whole or in part - can find their way into the shredder without going through the marking and identification steps. This is not a marginal process: almost a third of the proteins in the cell include unfolded segments, including proteins important for cell function, which play control and signal transmission roles. In fact, a fifth of all cell proteins are broken down in this way. Ostensibly, this is an efficient "vacuum cleaner", which maintains order and cleanliness in the cabin. But how do you prevent him from pumping everything that comes his way? This question is complicated to investigate - because the two routes, the "bureaucratic" and the passive, exist side by side in the cell.
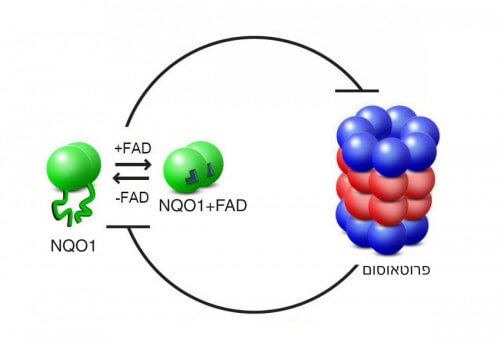
Several years ago, Prof. Yosef Shaul, head of the department of molecular genetics at the institute, discovered the first clue to the answer: he identified the "savior", which prevents the unwanted breakdown of proteins in the proteasome. It is an enzyme called NQO1, which leads a double life: it fights reactive oxygen compounds, thus protecting the cell from oxidative stress, and in addition, it also protects essential proteins from degradation. How exactly does he do this? This question was at the center of the current research, which was carried out by Oren Moskovitz, Nimrod Hazan, Hodia Kiser, Gili Ben Nissan, and Dr. Yitzhak Michalevsky from Dr. Sharon's group. To investigate in depth the interaction between the NQO1 enzyme and the proteasome in the passive degradation pathway, the scientists created an artificial experimental system, into which they inserted the enzyme and the "shredder" unit of the proteasome only - without the unit responsible for recognizing the ubiquitin label. This is how they managed to isolate and explore the desired route. The third factor that the scientists added to the system is a molecule derived from vitamin B2, called FAD. It is the binding of two FAD molecules to the enzyme consisting of two units of NQO1 that allows it to perform its functions in the cell.
Testing the NQO1 enzyme using mass spectrometry methods revealed that the binding of the FAD molecules stabilizes its spatial structure. When the scientists tested another form of the enzyme, in which the FAD binding site is defective, and when they removed the FAD molecules from a normal enzyme, a formless enzyme was created, unable to fold into an orderly structure. It was later discovered that these shapeless enzymes are broken down in the proteasome, and "disappear" from the experimental system. On the other hand, when the scientists added large amounts of FAD to the experimental system, they were able to restore the enzymes - the normal and the mutant - to their shape, thus saving them from degradation.
Research in Hayim cells, done in collaboration with Prof. Yosef Shaul and the research student from his group, Petar Tsavkov, expanded the picture. Administration of vitamin B2 to human cells led not only to an increase in the level of NQO1 and stabilization of its structure, but also to an increase in the level of p53. A similar picture was obtained by examining breast cancer cells, which do not contain the enzyme due to the defect in FAD binding. Adding B2 to these cells was able to rescue NQO1, thus allowing it to rescue p53.
The research findings, recently published in the scientific journal Molecular Cell, reveal that the interaction between the proteasome and NQO1 is based on mutual inhibition: the proteasome breaks down unstructured NQO1 enzymes, while the enzyme "blocks with its body" the destructive activity of the proteasome, and in this way saves from degradation not only itself, but also other proteins. An important component in this mutual control is an external nutritional factor - vitamin B2, which affects the structure of the enzyme, and thus controls its function. The scientists believe that it is possible that the phenomenon they discovered, that is, that a metabolic factor affects the function of a system through an effect on the structure, is not unique to the route they studied.
The mutation that impairs the ability of NQO1 to bind FAD, whose prevalence among white people is about 4%, exists in a fifth of the Asian population. The mutation means that the cells are unable to deal well with oxidative stress, and contain low levels of p53 and p73, which is why they are vulnerable to cancer - especially breast cancer and leukemia. The research findings hint at the nutritional importance of vitamin B2, especially in people suffering from the mutation.
The interaction between the proteasome and NQO1 is based on mutual inhibition

One response
How does NQO1 "know" how to save the desired proteins and not save those whose time has come? Thanks.