Over 900 additional human proteins that may bind to metal ions. The research promises to deepen our understanding of biological processes at the molecular level, as well as promote the development of innovative drugs
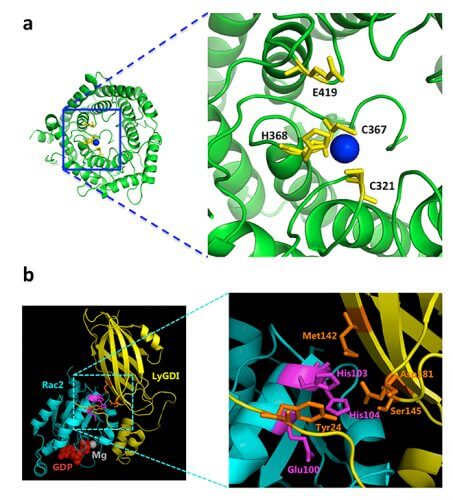
In science fiction stories, metallic body parts are often found, but our real body also depends on metal as an essential element. Electrically charged metal ions play key roles in enzymatic reactions, providing structural support to proteins in all tissues. In a new study carried out by scientists at the Weizmann Institute of Science, the scientists identified over 900 additional human proteins that may bind to metal ions. The research promises to deepen our understanding of biological processes at the molecular level, as well as promote the development of innovative drugs.
One of the main goals in biological research is to reveal the properties of proteins whose mechanism of action is unknown. There are about 20,000 proteins in the human body, but the first step in deciphering the three-dimensional structure - that is, creating crystals - has only been completed for a few thousand of them. As for most of the remaining proteins, all scientists have is the DNA sequence. But in the absence of information about their structure, scientists know little about their function.

About two decades ago, Dr. Vladimir Sovolov, from the group of Prof. Meir Edelman At the Weizmann Institute of Science, an algorithm that automatically describes the structural properties of a protein that interacts with small molecules, such as metal ions. Over the years, Prof. Edelman's group - which included Dr. Eran Eyal, Dr. Mariana Babor and Dr. Ronan Levy, then students at the institute - developed a computer algorithm that allows to predict with a high degree of accuracy, based on the DNA sequence, the The existence of metal binding sites in proteins, and the structures of these sites. In the new study, which Recently published in the scientific journal Proteins, Prof. Edelman and Dr. Sovolov used this algorithm to predict which proteins in the human genome might bind to metal ions. Research student Ariel Azia and Prof. Ron Ungar from the Faculty of Life Sciences of Bar-Ilan University, and Dr. Ronan Levy from the Institute's Department of Plant and Environmental Sciences participated in the study.
The binding to metal ions is one of the first characteristics that are checked when predicting the activity of proteins. This is due to the fact that, according to estimates, more than a third of all proteins are bound in part of their activity to metal ions, and this binding plays an extremely important role in various biochemical processes. For example, zinc ions, which bind to about a tenth of all proteins in the human body, are essential for the proper functioning of the nervous system and the immune system.

In the new study, the scientists used their algorithm to scan the DNA sequences of about 11,000 proteins, that is, more than half of all the proteins in the human body. The structure of most of these proteins is unknown, and it is often not clear what their role is in the body. From these, the scientists identified about 900 new proteins that, according to the algorithm, should bind to metal ions. Furthermore, the scientists identified the amino acids - which are the building blocks of proteins - that make up the possible binding sites in these proteins. Other scientists can now take advantage of the information discovered to discover the roles performed by these proteins and the amino acids involved in the process.
In the future, it may be possible to take the same approach not only in the context of the proteins encoded in the human genome, but also in relation to those of animals, plants or bacteria. About 100,000 genomes of different species, belonging to many kingdoms of life, will probably be sequenced by the end of this decade. Understanding the role of all the proteins encoded by these genomes is a huge task, and the ability to predict their properties, and especially their binding to the metal ions, can greatly help in this major task.
In the nearer future, the informed assessment that certain proteins will interact with metal ions may yield new insights regarding the mechanisms of various diseases. In a separate study, published in the scientific journal Human Mutation, the institute's scientists found that tiny genetic changes associated with many diseases often occur precisely in the DNA segments that encode the metal binding sites. The scientists identified 126 such diseases, including certain types of prostate cancer, cystic fibrosis, and sudden infant death syndrome. In 22 of these the connection to the metal ions was not known until now. The prediction that the metal binding site will be involved in the defect leading to the disease may help in the development of drugs designed to correct the defect.

