will enable the design of new drugs and improve the efficiency of antibodies
Proteins - the molecular "machines" that activate most life activities in biological systems - are created in a process that begins with a genetic sequence that encodes their structure, and ends with the folding of the protein chain and the creation of a precise three-dimensional structure that is essential for the protein's activity. A mistake or a change in the genetic sequence may result in the resulting structure not being able to properly perform the "role" that nature testified to it. It is a very complex system that was created and perfected over billions of years of evolution. From this it is possible to understand why the design of artificial proteins was considered for many years as a goal that "we may be able to achieve in the future". However, a new study by Weizmann Institute scientists SIt was published yesterday בProceedings of the National Academy of Sciences of the USA (PNAS) may bring the future closer to us.
Dr Sheral Fleishman, research students Dror Baran, Gabriela Pashula, Gideon Lapidot, and other members of Dr. Fleishman's research group in the Department of Biomolecular Sciences developed a computerized method that allows them to create artificial protein structures (for example, enzymes or antibodies), including those that do not exist in nature. For example, if they want to produce an antibody or enzyme with a certain physical structure, with the goal that it will bind to another structure, or perform a defined chemical action, they calculate the exact genetic sequence that will lead to the formation of a suitable protein chain - which will fold and create the desired three-dimensional structure. However, in repeated experiments it became clear that these "designed" enzymes or antibodies do not function. Like Hanukkah candles, these structures were also "for their viewing only".
What is there in a natural enzyme or antibody that is not in the same artificially created enzyme or antibody? Why do two structures with the same chemical composition, and with the same physical characteristics, differ so much from each other in everything related to chemical and biological function?"
Here, more or less, the research student Gideon Lapidot comes into the picture. He asked himself, and his teammates: what is there in a natural antibody that is not in the same antibody designed on a computer? Why do two structures with similar chemical composition differ so much from each other when it comes to chemical and biological function?
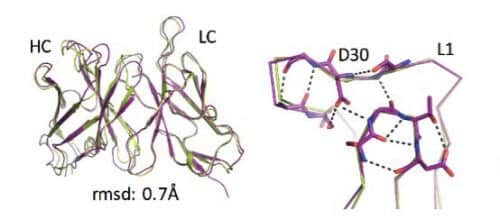
He decided to focus on "loops", those areas of the antibody molecule, which have a complex and non-repeating structure. It turns out that those unique loops are located in the "recognition areas" of the antibodies, and in many enzymes, in areas where they act on different molecules. That is, considerable parts of the structures of antibodies and enzymes play a relatively simple physical role, while it is the loops, the elusive components that are located at their ends, that perform the complex roles - "knowing the target" in the case of the antibody, and performing the cutting, or coupling of other molecules, in the case of the enzyme.
From this came an understanding that in order to create a functioning enzyme or antibody, it is not necessary to build them from scratch. Instead, it is better to distinguish between the main thing to treat. Between buildings whose main function is physical, and parts where the uniqueness and ability to operate lies. The scientists decided to create many different compositions of parts or segments that exist in nature - and thus also design the "loops" and the activity of the protein, or enzyme, or antibody. This, apparently, was the way of evolution to create so many types of enzymes, antibodies and other proteins that exist in nature today.
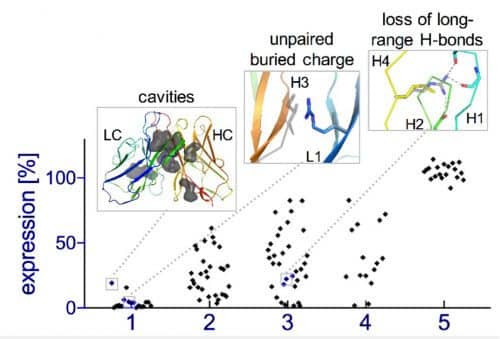
But evolution is a rather slow process. Development of a new family of antibodies, for example, can take millions of years. Because of this, the scientists returned to the route of designing protein structures in an artificial way. Equipped with their insights, this time they avoided attempts to recalculate the protein structures, and began the search for the appropriate structure from existing segments in nature, which they entered into a mathematical model that ran millions of different possible combinations. From the calculations, the researchers selected dozens of different designs of antibodies, and tested them again in the laboratory in an attempt to understand the reasons why some antibodies fold and function properly while others fail. After five rounds of trial and error, the researchers arrived at a computerized method that designs antibodies that all fold correctly in the experiment. In fact, a kind of evolution of the algorithm that calculated the protein structures took place here, with the experimental results "challenging" the algorithm and pulling it up the evolutionary ladder.
The artificial antibodies created in this way were stable, characterized by structural precision at the level of the single atom, and in addition, were active and attached to the target molecule very efficiently (in the experiments, the target that the antibodies aimed at was the insulin molecule).
In future experiments, the scientists plan to try and create artificial antibodies based on the model of antibodies from camels and llamas. It turns out that an antibody from the body of humans or many common animals is made up of about 200 amino acids. While many of the antibodies from the bodies of camels and llamas - animals that exist in harsh conditions - are made up of only about 100 amino acids. This fact may allow even greater precision in the design and production of artificial antibodies that may have many uses in the fields of research, medicine and industry.
