Researchers from the Weizmann Institute provide new information on the three-dimensional differences between the two types of aggregates of the p53 protein: the gap between the linker sequences causes the two pairs of p53 to rotate relative to each other
Its cause is through direct control over the expression of genes: when the cell is in a state of distress that requires a response, such as exposure to toxins or radiation, p53 is activated and binds to specific target sites on the DNA - that is, to a certain area of the genetic sequence. This way he can control the rate at which these genes will turn into active proteins, and in fact, he can determine the cell's response to the emergency.
"One of the great mysteries is how proteins like p53 manage to identify their target sites - a short sequence of bases - in DNA molecules that are three billion bases long, and to do so accurately and in a very short time," says Prof. Tzipi Shaked, Head The department of structural biology in the faculty of chemistry at the institute, which uses crystallographic methods to investigate the three-dimensional structures created by p53 when it binds to its target sites on DNA.
In the case of p53, it is a double mystery, since this protein knows a large number of target sites, each of which has a special DNA sequence, because it is required to regulate the activity of many genes. In addition to this, when the cell enters distress, p53 stands at a crossroads where it is required to "choose" between two main directions of action: one direction, in the case of relatively minor DNA damage, is to stop the processes of cell division and activate the mechanisms responsible for repairing the damage. A second direction, in cases where the genetic damage cannot be repaired, is the activation of genes that carry out the cellular suicide program (apoptosis), in order to maintain the integrity of the entire production. In each of the two options, p53 activates different genes, by binding to special target sites, each of which generates a sequence of events that activates proteins that handle the situation in different ways. How is the choice made between the two options? A possible clue to the solution of this question may be found in the way p53 binds to the various target sites. Many studies have shown that these sites consist of two binding sequences (10 base pairs long each), to which four p53 molecules bind. In 2006, Prof. Shaked's research group succeeded for the first time in creating crystals of p53 quartets bound to different DNA sequences, and deciphering their three-dimensional structures.
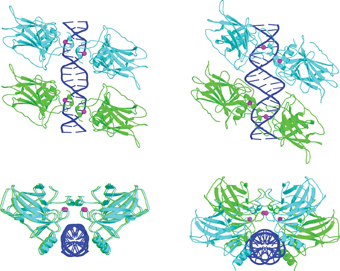
The research findings, published in the journal Molecular Cell, showed that two pairs of p53 proteins bind to the two DNA sequences next to each other, forming a quadruple complex. It was also discovered that target sites from different sequences have different patterns of interactions with p53: the amino acids found in the binding region of the protein rotate on their axis according to the DNA sequence to which they bind, in order to stick to it in the strongest way. In subsequent studies, in additional research groups, it became clear that although in all cases four protein molecules bind to DNA, the aggregates can be divided into two main types, according to their target sites: in the first type, the two binding sequences are separated by a sequence of pairs Bases of irregular length. In the second type there is no separation, and the two sequences are adjacent to each other. It was also found that sites of the second type, the attached one, are located before genes that are related to the arrest of division, while sites of the first type, separated, are located before genes that carry out the apoptosis program.
In the crystalline structures studied by Prof. Shaked's group in 2006, the target sites were of the separated type, so several questions were raised: Are there differences in the spatial arrangement of p53 clusters with the different target sites? If so, what do these differences mean, and how do they affect the interaction between the proteins and the DNA? And do they have a role in deciding to which pathway the damaged cell should be directed? Prof. Shaked and the members of her research group - post-doctoral researcher Dr. Malka Kiteiner, research associate Dr. Chaim Rosenberg, research student Oded Suad, and Prof. Dov Rabinowitz - studied a new type of crystals, in which p53 is bound to target sites of the tight type. Their findings, recently published in the journal Nature structural and molecular biology, provide new information about the three-dimensional differences between the two types of aggregates: the space between the linker sequences causes the two p53 pairs to rotate relative to each other.

"This seemingly small change leads to a complete change in the architecture of the aggregate, in the protein's affinity for DNA, and in the ability of additional proteins to join the structure. In fact, it's a world of difference," says Prof. Shaked. While on the target site of the contiguous type the quartet of proteins is arranged in two parallel pairs, in a planar parallelogram, on the target site of the non-contiguous type they form a twisted parallelogram (see figure). In the close form, the interactions between the four proteins are much stronger, the structure is much more stable, and the affinity for DNA is stronger. When the affinity is weaker, additional proteins are required to stabilize the structure. It is possible that additional such proteins are involved in the decision whether to try to repair damage, or to destroy the cell.
When the scientists examined the base pairs one by one, they discovered another interesting difference between the two target sites. It turns out that in the sites lacking the gain, the DNA bases of the adenine type are rotated 180 degrees with respect to their partners from the parallel strand, the thymine bases, creating a special structure. Calculations made by scientists from Columbia University in New York showed that a strong electric attraction contributes to the greater stability of structures of this type. "This discovery expands the known reading possibilities of the DNA sequence - not only in relation to p53, but also regarding other similar proteins," says Prof. Shaked. "It turns out that in addition to the information found in the sequence of bases, there is another alphabet, which lies in the angle between the bases, which further expands the identification possibilities of the genetic code."
The structural biology group of p53 also includes Dr. Naama Kessler, Dr. Yael Diskin-Pozner, Amir Alder, Aviva Kapitkovsky and Yaakov Halfon.

2 תגובות
First of all it was checked.
Second thing, since proteins have a limited lifespan and they are constantly being formed and being released, even if an abnormal protein is formed, the damage it will cause is relatively small. On the other hand the DNA is the software and if it changes the damage is permanent.
We constantly talk about damage to DNA by poisons or radiation, which causes mutations and damage to DNA that needs repair or destruction.
But what about damage to the auxiliary proteins such as P53, even if it is formed normally, later damage to it from radiation or toxins causes damage as if the gene that created it was damaged and then it does not fulfill its function properly, in my opinion it is enough for one or several proteins that do not do their function properly to to create malignant cells not as a result of a defect in the genes that create them.
It is true that we are dealing here with availability for damage, proteins that change quickly compared to DNA which is constant, but this aspect should still be checked as well.