Researchers in the faculties of biomedical engineering and electrical engineering at the Technion have developed a method for biological imaging in super-resolution with unprecedented efficiency

A group of researchers from the Technion has developed an innovative technology for biological imaging based on single molecules. The research was carried out by the student Elias Naama and the postdoctoral student Dr. Lucian Weiss under the guidance of Dr. Yoav Shechtman from the Faculty of Biomedical Engineering and Dr. Tomer Michaeli from the Faculty of Electrical Engineering at Viterbi. The new technology provides an accurate image with high resolution and unprecedented speed, without the need for prior knowledge of the shape of the model being tested. In an article recently published in the journal Optica, the members of the group present an innovative approach that leads to unprecedented reconstruction in terms of speed and accuracy (super-resolution). This is through deep learning in artificial layered neural networks.
Layered neural networks are systems that perform fast, efficient and accurate cataloging of data. Similar to the human brain, they are made up of layers of artificial neurons (nerve cells). This hierarchical structure allows them to analyze complex information and especially to identify patterns in this information.
The new technology developed at the Technion succeeds, through such networks, in producing a complete and accurate image directly from the raw information - information that comes from the light emitted from the studied model following its laser irradiation. In addition, it does not require any special skill from the user and does not require any prior knowledge regarding the shape of the model - this is in contrast to previous methods.
Resolution, or image sharpness, is one of the biggest challenges in optics. In traditional optical microscopy, it is limited by the "diffraction limit" - a limit formulated by the German physicist Ernst Carl Abe in 1873. Abe proved that the potential resolution of a microscope could not exceed a certain limit - approximately half the length of the light wave. In other words, if we use a wavelength visible to the human eye, we cannot separate objects smaller than ~200-300 nm. When it comes to tiny biological structures, this resolution is insufficient.
Since the 19th century, optical microscopy has been perfected, and new technologies have been able to "bypass" Abbe's law and produce much higher resolutions, called "super-resolution". However, microscopy of tiny biological structures is still a great challenge. This is partly because short waves, which enable high resolution, carry high energy and therefore may damage the biological cell. Furthermore, when we study living cells, the scanning speed is critical because of the dynamism of the living cell.
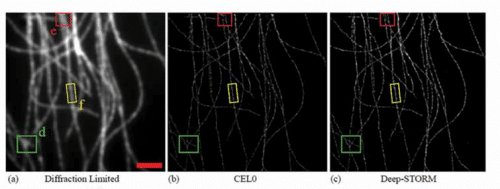
Positional microscopy, known by the nicknames PALM and STORM, is a new technology based on the blinking of individual molecules far from each other - far enough away that it is possible to determine the position of each of them. The information obtained is actually a sequence of images that are analyzed by computerized means and translated into one super-resolution image - a resolution improved approximately 10 times that of normal microscopy. The method earned its inventor, Eric Betzig, the Nobel Prize in Chemistry in 2014, along with Morner and Stefan Hell.
However, even this new technology suffers from problems related to the process of reconstructing the one image from the many flashes recorded. For example, when nearby molecules blink at the same time, their image overlaps and makes it difficult to identify their individual positions. In order to solve this problem, various computational methods have been developed over the years, but they suffer from high computational complexity, a very long running time and the need to select appropriate parameters that make it difficult for end users who are not experts in the intricacies of the algorithms to use. Therefore, the article by the Technion researchers is a very significant breakthrough in the field of microscopy.
The research was conducted with the support of the Google Research Foundation, the Zuckerman Foundation, the Technion (through the Career Advancement Chairship, the Ohlendorf Foundation and the Taub Foundation), the Alon Foundation and the National Science Foundation. The NVIDIA company donated the innovative Titan Xp GPU graphics card to the research group.
for the scientific article

3 תגובות
The son Yoav is the son of Dan Shechtman in my opinion.
The article has already been cited by 8 articles found in Arxiv, meaning they have not yet been published.
It does not have mathematics, it has application identification and matching the network to the application. which is not bad at all.
The articles about Binah are beautiful. Good articles.
In my opinion, there is only room for articles on basic research - Langland's bridging theories - a transition between theories for interdisciplinary application, Naftali Tashvi's information bottleneck theory - how artificial intelligence thinks, Verlind's theory of modified gravity, more precisely gravitational entropy - full professor in Zern and Utrecht, Holland . Former Princeton Associate Professor. Manages to calculate all dark matter effects without dark matter and move from one theory to another easily.