New information about signaling proteins, known as G proteins, may turn out to be extremely important in the fight against diseases such as heart defects and neurodegeneration and cancer.
New information about signaling proteins, called G proteins (The entry in Wikipedia), may turn out to be extremely important in the fight against diseases such as heart defects and neurodegeneration and cancer. For many decades, scientists have been wondering "how are signaling proteins transported and organized in defined complexes in the cell?". Researchers in the fields of nanoscience are now providing new information to solve this issue.
"We are now beginning to understand how signaling proteins are recognized and transported to specific areas of the cell and to gain a clearer understanding of the mechanism of important cellular processes such as cell signaling and growth. This important knowledge could be used in the future to understand diseases such as depression and Alzheimer's and cure them", explains Professor Dimitrios Stamou, who led this research.
Cells in the body are completely dependent on their selective ability to move and localize proteins to specific areas. Previous hypotheses, which suggested that proteins move in cells by recognizing nano-components in the membrane surrounding them, also known as fat collection, are now in fierce controversy. However, researchers from the Center for Nanoscience at the University of Copenhagen discovered an unknown mechanism based on the shape of the cell membrane and which was published in the scientific journal Nature Chemical Biology.
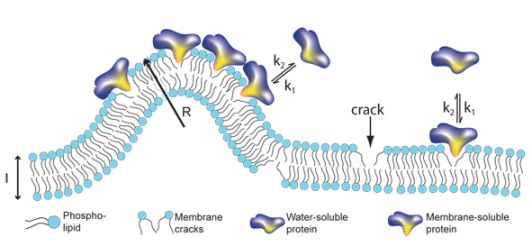
Like all other materials, cell membranes will crack as a result of their bending. However, the cell membrane has a unique property: bending more and more does not create larger cracks but a higher amount of cracks of the same size. It turns out that a number of important proteins "prefer" to bind to these fissures and thus the bent parts of the membrane become a good "meeting" place for them to join together to perform complex tasks that require the joint activity of many different proteins.
"We were surprised to find that the number of slits in the membrane determines the number of proteins that will bind. Until now, researchers in this field believed that the decisive factor was the ability of the proteins and their tendency to bind to the membrane, a concept known as affinity. Our findings contradict this hypothesis," notes one of the researchers.
The proteins move inside the cells in small vesicles - a kind of soap bubbles that, like the cells themselves, are surrounded by a shell. The researchers prepared vesicles of varying sizes in the laboratory and tested how many different types of proteins could bind to their membranes. They discovered that the smaller the size of the vesicle and the more bent the envelope, the higher the number of cracks and as a result more proteins, relative to a unit of area, can bind.
"As soon as we realized that the most decisive factor in our observations was the structure of the membrane, we immediately thought that we might have found a general mechanism that could also be adapted to other types of proteins besides the ones we studied. Following this, we next investigated G proteins which are important signaling proteins linked to the cell membrane in different ways by a lipid "anchor". Our findings verified that the mechanism is indeed general," explains one of the researchers.
"The discovery of the immeasurable importance of the membrane structure for the demarcation of hundreds of important signaling proteins is essential for our understanding of the multitude of biological processes, many of which are directly related to common diseases," emphasizes the lead researcher.
