How do we know how to locate our nose in space or follow the position of the arm as it moves without the sense of sight? Scientists from the Weizmann Institute have revealed some of the molecular mechanisms responsible for the proprioception system - self-sensing of the position of our body organs and their movement.

Close your eyes and try to touch your nose with your index finger. Most of us are able to perform this movement with great precision, but how do we know how to locate our nose in space without the sense of sight? And how do we manage to track the position of the arm as it moves? "Few people know that in addition to our five senses - sight, hearing, smell, touch and taste - there is also what is sometimes called the 'sixth sense' - the sense of location in space," says Prof. Yoram Groner, from the Department of Molecular Genetics at the Weizmann Institute of Science. Such bodily awareness, also known as proprioception, refers to the self-sensing of the position of our body parts and their movement.
When there are neurological disorders, the proprioception system is often affected, therefore, a simple "finger-to-nose" test is used by doctors to initially diagnose these disorders or to rule them out. However, even though proprioception is one of the simplest neural "wiring" systems in the body, the molecular mechanisms responsible for creating the sense of position in space are still not well understood.
In a study that lasted for several years in Prof. Groner's laboratory, full faculty scientist Dr. Ditza Lebanon and her colleagues discovered that a certain protein - the Runx3 transcription factor - is an essential controller for the development and function of TrkC-type nerve cells (neurons), which are special sensory neurons of importance in proprioception. Dr. Lebanon explains: "We found that the loss of Runx3 activity is related to the death of TrkC cells in the ganglia on the sides of the spinal column in mice, and as a result they develop ataxia, meaning they lose the ability to place their legs in the right place and their ability to control their movement and body balance. Although the DNA segment that encodes Runx3 is present in all body cells, and probably also in other types of sensory neurons, the gene is 'activated' - that is, transcribed and translated into a protein - only in TrkC type sensory cells, and the question we are trying to answer is, what exactly? "turns on" and "turns off" the Runx3 gene at the right place and time, thus leading to the formation of this type of nerve cells."
Few people know that in addition to our five senses - sight, hearing, smell, touch and taste - there is also what is sometimes called the 'sixth sense' - the sense of location in space"

Dr. Levanon and her colleagues - Dr. Elena Appel, Dr. Kira Orlovsky and Dr. Yehuda Salzberg - discovered three key players who manage the transcription of the Runx3 gene, and in this way are involved in the development and function of proprioception. In order to begin "gene transcription", the first step in the process in which the body turns the information encoded in the gene into a protein, transcription factors bind to a specific sequence in the DNA around the gene, which functions as an initiating segment (promoter) of the transcription process. But the control system that controls the transcription is complex, and includes additional control factors, so another problem arises: how to locate and where to look for the genetic controllers involved in the transcription of the Runx3 gene.
To deal with this difficulty, the scientists used bacterial artificial chromosomes (BACs), which contain large sections of murine DNA, and created genetically engineered mice that contained BACs carrying the Runx3 gene, including the genomic regions adjacent to it. In addition, to enable monitoring of Runx3 expression, a fluorescent reporter gene (GFP) was inserted into Runx3 in BACs, which was used as a gene activity detector. "Investigating the expression pattern of the reported gene in transgenic mice containing BACs, allowed us to discover a remote region which is responsible for the specific expression of Runx3 in TrkC type neurons, and which contains the control factors we were looking for", explains Prof. Gruner. Dr. Lebanon adds: "Since the expression of the Runx3 gene in TrkC neurons has been preserved throughout evolution, we believed that its control factors have also been preserved, and therefore we compared the section that includes the control factors that we identified in the analysis of the results obtained from transgenic BAC mice, to corresponding regions in the genomes of Bani Man, birds, frogs and fish. The comparison allowed us to identify three genomic segments in a distant region from the Runx3 gene, which were common to all of them." These three transcription controllers were named: R2, R1 and R3.
To prove unequivocally that these transcriptional controllers (R) indeed control the expression of Runx3, the scientists used an innovative research method that uses CRISPR/Cas9 to omit from the mouse genome each of the control regions, individually or in pairs. the findings, recently published in the journal Genes & Development, clearly show that deleting two of the three control factors – R1 and R3 – had the same effect as deleting the entire Runx3 gene itself; That is, mice that lacked these two short genetic segments developed ataxia.
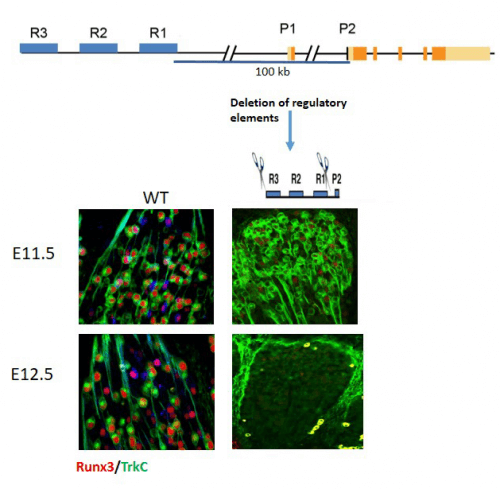
The continuation of the research revealed that for the specific development of TrkC type neurons it is not enough that the controllers direct the transcription of Runx3 and accelerate its expression in TrkC type neurons; At the same time, they also work to suppress its expression in other types of nerve cells. Furthermore, the researchers also insisted on the unique role of each controller, which led to the discovery and characterization of subpopulations of TrkC cells: in the early stages of mouse embryo development, in the middle of the 12th day, the three controllers - R2, R1 and R3 - operate together in managing the expression of Runx3 in neurons of the TrkC type, but with embryonic development, a complex mechanism of mutual effects between the three controllers begins to operate. This mechanism governs gene expression in TrkC type neurons and subtypes of the cells of the proprioceptive system.
"The importance of these results stems from the fact that there are only a handful of studies documenting the communication between transcription controllers and the genes they manage in the living body itself, and not in a test tube," says Dr. Lebanon. "In the future, it may be possible to use the three transcriptional controllers we discovered to target TrkC neurons in cases of ataxia and other neurological diseases in which Runx3 is involved."
