When the DNA splits into two separate strands, to allow for its repair or replication, the "single" strands are unstable, and their vulnerability to environmental damage increases. In a new study, Weizmann Institute of Science scientists presented a computational model that shows how the "support" proteins that stabilize the individual DNA strands work.
the question: How do the proteins that stabilize the single DNA strands work?
Findings: The institute's scientists presented a computational model that shows that the stabilizing proteins and strands wrap around each other, thus forming stable aggregates.
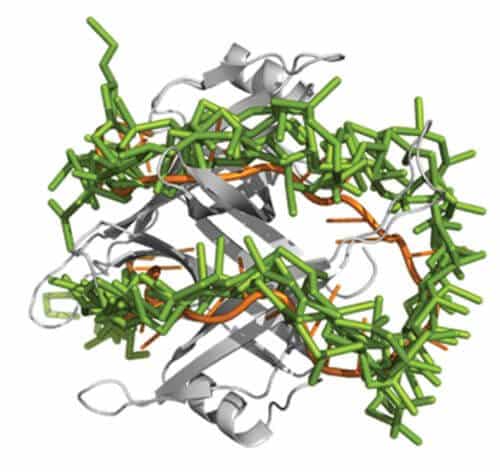
Everyone knows DNA as a double helix, but when it comes time to repair, replicate, or "read" the information inside, the DNA splits into two single strands. This fragmentation makes it accessible to "read" the genetic information contained in DNA, but as a result of their "solitude", the strands are unstable, and their vulnerability to environmental damage increases. In a new study - recently published in the scientific journal "Proceedings of the National Academy of Sciences of the United States of America" (PNAS) - Weizmann Institute of Science scientists presented a computational model that shows how the "support" proteins that stabilize the individual DNA strands work. These proteins protect the strands, preventing them from folding back into the double-stranded structure, which may make it difficult to access the information inside.

Similar to most of the proteins that come into contact with the double-stranded DNA as part of the search for the unique binding site, the "support" proteins slide over the single DNA strands. While surfing, the contacts between the proteins and the DNA are usually based on a difference in electrical charge: a positive charge for the proteins, and a negative charge for the DNA.
The model built by Prof. Jacob (Kubi) Levy and post-doctoral researcher Dr. Garima Mishra, from the Department of Structural Biology at the Weizmann Institute of Science, reveals that in the case of the support proteins - the situation is more complex. These proteins react with individual DNA helices also chemically, using amino acids that contain structures called aromatic groups. Through these two types of contacts, the proteins and individual strands wrap around each other, forming stable aggregates. The new model allowed the institute's scientists to predict the exact structure of these aggregates, and after comparing it to known structures discovered in experiments carried out on crystals of the molecules, it turned out that the prediction was correct.
During the surfing, the contacts between the proteins and the DNA are established
About differences in electric charge:
A positive charge for the proteins, and a negative charge for the DNA
The model serves as a very valuable tool for future studies on the interactions between the support proteins and the individual DNA strands. These relationships are a challenge in experiments, because they are more complex than the contact that exists between proteins and the DNA double helix. The complexity arises from the fact that the single strands are more flexible, and their chemical variation relative to the double helix is greater. The new model provides the theoretical information needed to facilitate single strand experiments.
Among other things, the model may allow scientists to check whether findings discovered in recent years - which concern the interrelationships between DNA double helices and proteins - are also valid for the contacts that exist between the "support" proteins and the individual strands. For example, the researches of Prof. Levy and the members of his group have previously contributed to the solution of the mystery - how do proteins that slide on the double helix manage to find binding points very quickly, and at the same time create bonds strong enough so that molecular contacts occur at these points. Through computational analysis, the institute's scientists identified structural and energetic characteristics that allow proteins to bind to DNA with maximum speed and strength. Among other things, the scientists showed that the proteins are able to "jump" over obstacles, and take advantage of structural data, such as their unfolded parts.
This research sheds new light on the function of DNA at the molecular level, thereby advancing our understanding of the biological processes that are at the basis of life. In addition, it is possible that it will lead to the development of biotechnological applications based on the contacts between proteins and DNA.
| Science books Computer simulations of approximate molecular models make it possible to predict in less than 12 hours the complex structure of a support protein "surfing" on single-stranded DNA. |
