Certain mutations, common in populations of African descent, greatly increase the risk of end-stage renal disease. Surprisingly, it was found that the same mutations also protect against the African sleeping sickness transmitted by the tsetse fly. Understanding the biological mechanism in which this gene is involved may advance the study of kidney diseases and at the same time, also the fight against African sleeping sickness
By: Shai Tzur and Karl Skortsky
African Americans and renal failure
End-stage renal failure, also known as kidney failure, is the last stage of chronic kidney disease - a name that refers to a wide variety of diseases that harm kidney function. At this stage, the functions of the kidney, which is responsible, among other things, for eliminating toxins from the body, are extremely poor. Patients suffering from end-stage renal failure need ongoing medical treatment with dialysis or a kidney transplant. The number of patients with end-stage renal failure in the United States today is about half a million, and about one hundred thousand new cases are diagnosed there every year. In Israel there are over 5,000 patients who need regular dialysis treatment. The main causes of chronic kidney disease are diabetes, hypertension, nephritis and many other diseases that damage the kidney. In the last decade, in which there was a significant increase in the rate of morbidity in the common metabolic diseases (diabetes, hypertension), there is also an increase of about 30% in the number of patients with kidney diseases. According to a common estimate, 13% of the residents of the United States suffer from one degree or another of chronic kidney disease, which can lead to end-stage renal failure. In addition to environmental factors that may increase the risk of end-stage renal failure (inaccessibility to medical care, exposure to toxins, etc.), there are Also evidence supporting the existence of genetic risk factors. Epidemiological data from the United States show that in the African-American population, the degree of risk of the disease is up to five times higher than in the population of European descent. One third of dialysis patients in the United States are African-American, even though they make up only one tenth of the population. Studies have even shown that family members of African-American patients have a higher risk of developing kidney disease compared to people whose families do not have patients. The socioeconomic status of African Americans is only a partial contribution to the high rate of kidney disease among them. Many findings thus indicate a common genetic factor involved in the onset of kidney diseases in African Americans. Complex diseases are diseases that originate from a combination of environmental and genetic factors. Diseases such as diabetes and hypertension, as well as kidney diseases, appear more frequently among people who lead a western lifestyle (environmental factors) and among those whose relatives have the disease (genetic factors). For the most part, the genetic factors have less influence on the appearance of complex diseases. On the other hand, classic genetic diseases (Mendelian, for example: Tai Sachs, Canavan, etc.) are directly caused by a single genetic factor and independent of environmental factors.
Following the statistical affinity
In studies designed to identify the genetic factors that influence the risk of contracting a complex disease, it is customary to use a genome-wide association study (GWAS), during which a genetic scan of the entire genome is carried out in a large number of participants. From a technical point of view, a chip is used that enables the identification of the instance of each of the hundreds of thousands of markers being tested, located at different sites in the genome. These markers are usually sites that have single nucleotide polymorphisms (SNPs) in the DNA. Diversity results from a historical change that occurred at the genetic site (mutation), which resulted in the addition of a new instance or allele that is now common in the population. These changes are mostly neutral and have no biological effect. For example, in a specific location on chromosome 22, in the MYH9 gene, the marker rs5750250 is located. In the genetic scan of the entire genome, this site was also examined and it was determined which of its alleles each subject carries. In the method of genome-wide association research, hundreds to tens of thousands of DNA samples of participants, divided into a patient group and a control group, must be scanned. In the analysis of the findings, the statistical affinity of each of the thousands of genetic markers tested for the disease under study is examined, by comparing the frequency of a particular allele in the patient group to its frequency in the control group, and locating markers in which the difference in frequency is statistically significant. Such a significant difference indicates the possibility that the marker found (or an unknown mutation near it) is responsible for a biological functional change that increases the risk of the disease being studied. Identifying a statistical relationship between a certain genetic marker and a genetic disease is only an initial step in research. Later, the genetic region where the marker was discovered was investigated in depth, through the characterization of additional markers in its environment and with the help of additional samples, and an attempt was made to locate the genetic variation that causes biological change and the appearance of the disease. In very few cases, a genetic factor with a strong affinity to the disease and a clear biological effect is identified. In these cases, the researchers strive to understand his involvement in the mechanism of the disease in the quest to find a cure for it.
Mistaken identity
Back to kidney disease in Africans. The high frequency of chronic kidney disease in African Americans compared to populations of European origin indicates, therefore, that some genetic factor is involved in the disease. But it also indicates another thing: that this genetic factor should be found with high frequency in the African-American population and be relatively rare in people of European origin. This fact allowed the use of a unique method for genetic scanning based on the genetic difference between different populations, known as admixture mapping. This method is a derivative of the method described above (GWAS), and its great advantage is economic: with it, the number of markers tested in the genome can be greatly reduced, and genetic characterization of about 2,000 markers throughout the genome is sufficient to obtain a result (instead of 500 markers in a "normal" genetic scan ). Such a genomic scan can only be performed in a mixed population that originates from the mixing of source populations with a very different genetic background, and very different from each other in the prevalence of the disease being studied. Therefore, this scanning method suited our example, where the frequency of the disease is high in Africans compared to Europeans, and at the same time, the genetic background of the populations is very different. And finally, we have at our disposal a population of mixed people - Afro-Americans. Surprisingly, it became clear in recent years that the genome of the members of this seemingly homogeneous population is about 20% European. In other words, on average, a fifth of the genes of an African-American person are "white" European. The significant component of European DNA in African-Americans is the result of mixing between the populations during the past hundreds of years when they lived side by side in America, and also results from the rape of black women during slavery. Since the presumed genetic factor involved in kidney disease is of African origin, it was possible to search for and locate a common region in the genome of the patients rich in markers of African origin through the genetic mapping method of mixed race, which was carried out in a sample of non-diabetic African-American patients. This region is supposed to contain the genetic cause of the disease, and about two years ago researchers Jeffrey Kopp and Linda Kao discovered it at the same time. The extensive region, located on chromosome 22, contains dozens of genes, including the MYH9 gene. This gene attracted the attention of researchers as a prominent possible candidate to be the cause of the disease, as it is known that mutations in its axons cause a syndrome whose symptoms are kidney damage. Today it can be said that this determination was premature and not sufficiently based, as will be described later. In the last two years, dozens of follow-up studies on the MYH9 gene have been published, but none of them have been able to identify what exactly goes wrong in its activity, which could increase the risk of kidney disease.
Read by numbers
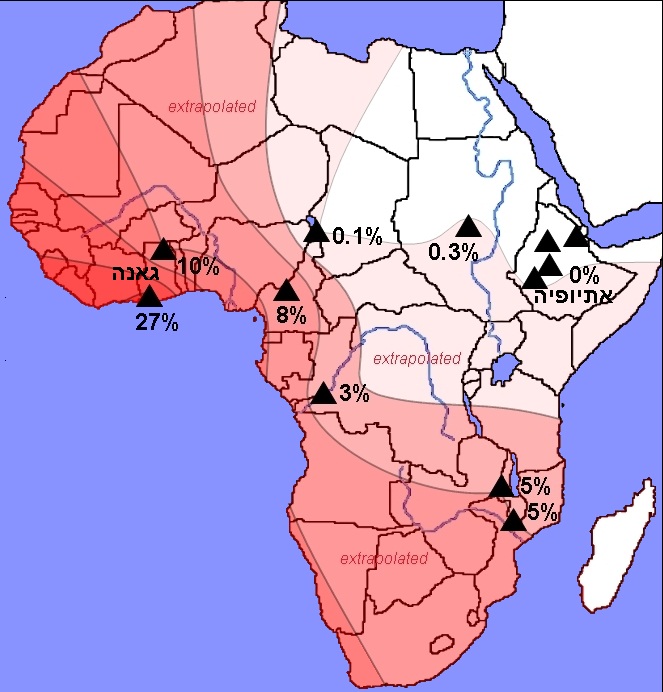
The focus on the MYH9 gene ended when preliminary information obtained in the "1,000 Genomes Project" was published. This is a revolutionary international project, which aims to determine the complete genetic sequence of at least 2,500 people from populations around the world. Following the technological developments in the fields of genetics, robotics and computing, today it is possible to determine the complete genetic sequence of a single person within a few months and at a cost of only tens of thousands of dollars. For comparison, the human genome project, at the end of which the complete genetic sequence of one and only genome was published, took 13 years and cost a huge financial investment of three billion dollars. At the beginning of 2010, as part of the project, the complete genetic sequences of approximately 120 people were published: 60 Africans from Nigeria and 60 Europeans. The newly published information was used by us to locate genetic factors involved in kidney diseases. Our group, which consisted of researchers from the Faculty of Medicine at the Technion, Rambam Hospital, Tel Aviv University and Hadassah Hospital in Jerusalem, and also included researchers from abroad. The group was led by Prof. Karl Skortsky, Dr. Sahron Rost, Dr. Tali Shemer, Mr. Gennady Yudukovski, Dr. Sara Zelig, Dr. Walter Wasser, Dr. Doron Behar and the doctoral student Shai Tzur. Our group analyzed the new information published following the 1,000 Genomes Project, and quickly identified genetic sites (mutations) outside of the MYH9 gene that were previously unknown and not suspected of being involved in kidney disease in African Americans. The study (Tzur et al.) was published in the journal Human Genetics in July 2010. After identifying the suspicious mutations in a nearby gene called APOL1, their prevalence was tested in a sample of 955 DNA samples. The sample included samples from non-diabetic end-stage renal failure patients, as well as from people whose kidney function is normal. The samples were sourced from blood tests collected from participants of African American or Hispanic American descent who visited dialysis clinics in New York, United States. The analysis of the findings revealed a very strong relationship between the mutations being tested and end-stage renal failure. The affinity is particularly high in a recessive (homozygous) model, that is, the risk of the disease is highest when a person carries two dangerous alleles at the same time (in West Africa about 10-25% of the population are homozygous). The new mutations we identified corresponded to the two main conditions we were looking for: high frequency of the risky allele in Africans and its rarity in Europeans; And a functional effect is expected on the protein activity due to causing a change in the amino acid sequence. We found that the affinity for the disease of the mutations in the APOL1 gene (S342G, I384M) is much higher than the affinity of the markers previously found in the nearby MYH9 gene, which the scientific community focused on in the two years prior to our study. The mutations we identified in the APOL1 gene are the most common genetic factor with the strongest link ever found to any complex disease. In addition to the genetic characterization of the mutations in the patient sample, we also performed genetic characterization of samples from 12 different populations in Africa. The samples were collected by our colleagues in London, Dr. Neil Bradman (Bradman) and Dr. Ayala Tarken (Tarekegn), and in Ethiopia by Prof. Andesho Bekele (Bekele). About 670 DNA samples were used to determine the frequency of the risky allele in each population. We found that the frequency of the poor allele is very high in central and western Africa, especially in the humid tropical regions, inhabited by populations speaking Bantu languages. On the other hand, in areas with a dry climate on the border of the Sahara desert in the north and Ethiopia in the east, we found that the frequency of the allele is zero (see figure).
This finding is interesting for two reasons: a. It is known that in Ethiopians the frequency of terminal kidney failure is extremely low. Therefore, the absence of the risky alleles of the APOL1 gene in this population strengthened the hypothesis that they increase the risk of the disease. B. The correlation we found between the geographic distribution of the mutation and a certain type of climate and geographic environment suggests the possibility of a strong influence of natural selection. Many studies have shown that adaptation to environmental factors in Africa, and in particular to pathogens, had a great impact on shaping the human genome. Here, too, it was found that natural selection influenced the appearance of mutations in APOL1 and the increase in the risk of kidney diseases, as will be described below.

4 תגובות
Indeed yes, later on the issue of selection is explained as a result of environmental pressure that results from the hoverfly that causes the African sleeping sickness.
The mutations that protect people who carry them from sleeping sickness, increase the risk of kidney disease. A similar story to sickle cell anemia in Africa, and protection from malaria.
Too bad the article is incomplete here.
For 1 and 2: The answer is probably found later in the article on the Bioinform website.
Agree with Naomi
It's ridiculous that the only time the word "fly" appears on this page is in the title.