It is an amino acid that the body produces but was discovered to science only in 1986
With a few exceptions, all proteins known to us consist of only twenty amino acids. 25 years ago scientists discovered the twenty-first acid - selenocysteine, and a decade ago the twenty-second acid - pyrrolizine. However, the mechanism by which the cell produces these rare building blocks has remained unknown until now. Now, researchers from Germany have managed to reveal the structure of an important enzyme essential for the production of the amino acid pyrrolizine.
Proteins play key roles in many vital processes in living things: they transport substances from one place to another, catalyze chemical reactions, "pump" ions or recognize signaling molecules. The complexity and variety of the proteins are great, when in the human body alone there are more than a hundred thousand different proteins. And yet, almost all of them consist of only twenty amino acids. Only a few proteins with a particularly high level of expertise also contain the rare twenty-first amino acid, selenocysteine, which was discovered only in 1986.
A big surprise was also the discovery of the twenty-second amino acid in 2002 in ancient methane-producing bacteria (or archaeons): pyrrolysine. This acid is genetically encoded in a similar way to the other amino acids. These bacteria use this rare amino acid for proteins they require for energy conversion processes. Pyrrolysine is located in the catalytic center of the proteins and is essential for their function - the energy production process in these bacteria will not occur without this amino acid.
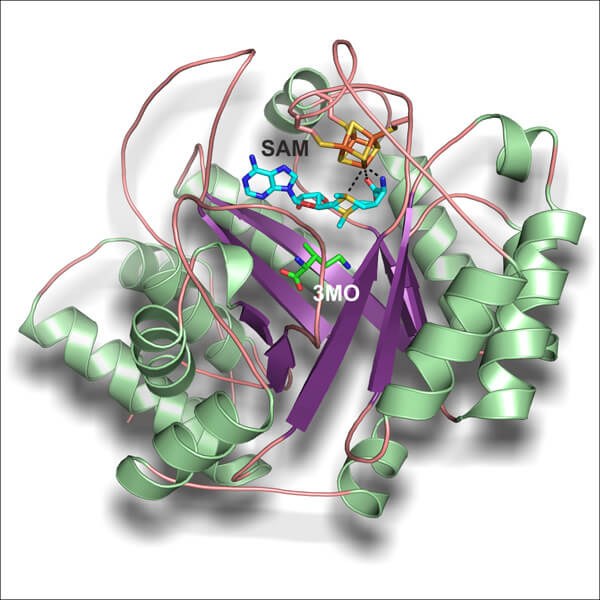
In March 2011, scientists from Ohio University were able to decipher parts of the pyrolysin production process. They proposed a reaction mechanism in which a specific enzyme catalyzes the first step of pyrolysine biosynthesis by converting the amino acid lysine to the intermediate methychlornithine. In the next step, the researchers were able to determine the crystal structure of this enzyme using X-ray analysis.
To their astonishment, they were able to observe the enzyme at the "time of action" itself: at the moment of crystallization of the reaction product, metychlornithine, when it had not yet been released from the listeners. "The situation in which the product was still encased in the enzyme is a special and extremely discounted find," says one of the researchers. "Not only were we able to directly identify the intermediate product, but we were able to retrospectively explain the formation of this substance from the original amino acid lysine."
Not only was this reaction previously unknown, but it was very difficult to speed it up. An accumulation of four iron atoms and four sulfur atoms in the active site is the basis of the conversion process. "This is truly an unusual catalytic reaction. Until now, no chemist in any laboratory has been able to synthesize the intermediate (methylornithine) in a one-step reaction from lysine," explains the lead researcher.
Understanding the conversion process of lysine to methylornithine helps scientists understand how ancient bacteria can adapt an existing system to make a new and needed amino acid, when it is inserted into a suitable protein catalyzing a very specific reaction. The researchers will be able to use this new knowledge in order to create artificial amino acids in order to create personalized enzymes with special properties, which could, for example, be used in a variety of applications in the fields of biotechnology and medicine.
The news about the study

2 תגובות
The genetic code is the same for all creatures. The pyrrolizine is encoded by the stop codon, UAG.
In organisms that are not supposed to use pyrolysin, i.e. those that do not produce methane, the enzyme needed to create pyrolysin is missing, and the UAG is called a stop codon. When the ribosome reaches it, peptide synthesis stops.
In archaea that produce methane there is the enzyme that produces pyrolysin, therefore there is also tRNA that is related to pyrolysin. When their ribosome reaches the UAG found with a nucleotide sequence that "supports" the insertion of pyrrolysin on the mRNA, it will attach the pyrrolysin to the peptide that it synthesizes.
A check in the genetic code table shows that the amino acid pyrrolizine is not coded, contrary to what is written here:http://en.wikipedia.org/wiki/Genetic_code
Does this mean that its genetic coding appears only in those bacteria, or in those bacteria there is simply a later conversion of lysine regardless of the genetic coding?