The institute's scientists designed an artificial enzyme, put it through a process of evolution in a test tube, and improved its performance

In the close competition that exists between nature and man, the scientists recently won several important points, when they managed to create, out of nothing, a new enzyme, of a type that does not exist in nature. This achievement opens the door to a variety of future applications, in the fields of medicine and industry. Enzymes are, without a doubt, a striking example of the achievements of evolution. These molecular machines, without which life would not be possible, carry out all the chemical processes that take place in the living body. Millions of years of natural selection have perfected and improved their activity, so that they are able to speed up the rate of certain chemical reactions by billions and even more. In order to create a new enzyme, which does not exist in nature, a thorough understanding of the principles of action and the structure of the enzymes, and an advanced ability in the field of protein engineering is required.
A team of scientists from the University of Washington in Seattle and the Weizmann Institute of Science succeeded in this task, for the first time in the world. The research findings were recently published in the journal Nature.
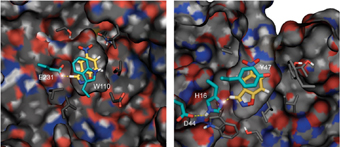
An enzyme is a biological catalyst. It is a protein molecule made of a "string" of amino acids, which fold to form a defined three-dimensional structure. The team of researchers approached, in the first step, to design the core of the machine - the "active site" - where the chemical reaction takes place. The role that the scientists attributed to the new enzyme is to remove a proton from a carbon atom. To this end, they created an array that includes an amino acid whose role is to "pick up" the proton, and additional amino acids that speed up the process of transferring the proton.
The next step in the process was designing the enzyme skeleton, that is, determining the sequence of the approximately 200 amino acids that make up the protein. Apparently, the number of ways to arrange amino acids from 20 different types, in a string consisting of 200 units, is almost infinite. But in fact, only a limited number of possibilities actually come into consideration, because the sequence of amino acids determines the spatial structure of the enzyme, and therefore also affects its activity. Prof. David Becker from the University of Washington in Seattle used computerized methods to scan tens of thousands of possible sequences, and located about 60 structures capable of supporting the designed active site. Eight of them successfully passed from the computer model stage to the protein stage in vitro, and were found to be biologically active. Of these, the two most effective enzymes advanced to the "final stage". Dr. Orly Diem-Boutbol and Dr. Shira Albeck, from the Department of Structural Biology at the Weizmann Institute of Science, deciphered the spatial structure of these enzyme molecules, and confirmed that the structures created in practice are almost completely identical to those designed and designed using the computer software.
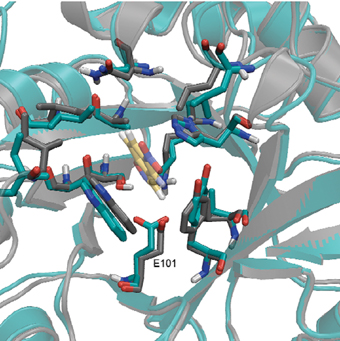
At this point, the efficiency of the new enzymes fell far short of the efficiency of natural enzymes perfected over millions of years of evolution. Here the ambitious research could have encountered a significant difficulty, except that Prof. Dan Toufik and his research student, Olga Khersonsky from the Department of Biological Chemistry at the Weizmann Institute, developed a method that allows protein molecules to go through a process of accelerated evolution, which mimics natural evolution. This method is based on causing random mutations, and scanning the variety of enzymes created, in order to find those that have improved their efficiency. These enzymes go through another round of mutations, and God forbid. Seven cycles of "evolution in vitro" improved the efficiency of the new enzymes 200 times compared to their original efficiency, allowing them to speed up the selected reaction a million times compared to a chemical reaction occurring without an enzyme.
Thus, for example, mutations in the shell region of the active site caused small spatial changes in the structure of the site, which corrected defects in the computerized planning of the position of the amino acids in the active site. It also turned out that the correction of tiny defects, at the levels of a millionth of a millimeter, greatly affected the rate of proton passage. Other mutations increased the flexibility of the enzyme, thus helping to release the product from the active site more quickly.
"The combination of technologies - determination of structure through computerized planning, and an evolutionary process in vitro - opens up new horizons in the production of artificial enzymes," says Prof. Toufik. "Thanks to this research, we better understand the structure of enzymes and their modes of action. This understanding will enable the design and production of enzymes for applications that nature did not think of, such as detoxification, drug production, and many other processes."

7 תגובות
He meant to remove a proton (H+) from a carbon atom in the chain
Right point.
I too was alarmed when I read this and then forgot to comment.
It sounds like alchemy.
Remove a proton from a carbon atom? enzyme? Please correct…
Wow Wow wow…! What an amazing creation! So important for the future
The order of the digits has been corrected. Thanks for the comment.
The molecular world simply amazes me every time. Kudos to the researchers.
To the editor of the article:
The order of the digits in the numbers has been reversed.
For all evolution fools:
So tell me please, how is it that even though there is no evolution we manage to use it in any field we want.
On second thought: it's better not to say.