Researchers from the University of San Diego have developed a method to reduce the time required to determine the structure of proteins obtained from natural compounds from a period of six months to a year to a period of only one day
Determining the exact structure of an unknown natural compound is a slow and expensive step in the screening and development of new drugs - however, this limitation will be removed in the near future thanks to a new combination of experimental and computational methods developed at the University of San Diego in California and presented at the RECOMB 2008 conference in Singapore.
Researchers from the University of San Diego have developed a method to reduce the time required to determine the structure of proteins obtained from natural compounds from a period of six months to a year to a period of only one day. This significant progress could help researchers in the field of drug discovery - who need as much and as fast information as possible regarding natural compounds with important medical properties such as activity against bacteria, viruses and other parasites being tested by them.
According to the researchers, today it is complex, expensive and time consuming to determine the molecular structure of a family of natural compounds called "nonribosomal proteins" (nonribosomal peptides, NRPs) which are being intensively tested to discover their medicinal properties. In order to help with this issue, the researchers developed a fast, automatic and cheap method for determining the structure of these proteins through an innovative collaboration between mass spectroscopy experts from the School of Pharmacy and bioinformatics and computer science experts from the School of Engineering.
If we describe the structure of these proteins as an annular chain of beads, then the new method can decipher both the exact mass of each of the separate beads based on mass spectroscopy and determine the order of the beads within the chain - two crucial pieces of information for deciphering the structure of the compound and its medicinal activity. In addition to the discovery of new drugs and the study of natural compounds, the authors claim that their research can also assist in the efforts of biotechnology engineers to reprogram sequences of Escherichia coli in order to transform them into an array of proteins of the above type, now researchers have a rapid method for characterizing proteins of this type. Such proteins, such as penicillin and other natural compounds, are the pinnacle of success in the pharmaceutical industry - nine of the twenty best-selling drugs today were produced from natural compounds, say the researchers.
Proteins of this type have evolved over millions of years and are usually used as chemical defense and internal communication factors in the various organisms, which are produced within them, as one of the researchers explains. The difficulty in determining the structure of these proteins is notorious and this is because the usual protein fitting methods do not work in the above case. Their ring structure, the presence of unusual amino acids (not found in common databases) and the lack of structural information found directly in the genomic DNA (due to the non-ribosomal nature of the proteins), all together constitute unique difficulties. Until today, researchers had to rely on slow, manual, expensive and unreliable methods to decipher the structure of these proteins. "This work removes a particularly troubling bottleneck in the field of drug discovery for this type of medicinal material," says one of the researchers. "We demonstrated a rapid method for characterizing non-ribosomal proteins. Our next step is to repeat these findings in the discovery of new proteins of medical importance."
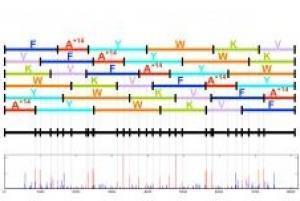
The researchers showed that these ring proteins can be unfolded into open chains and then the resulting peptide chains broken down into smaller and smaller subunits of the original ring using mass spectrometer graded steps. This approach - known as "multistage mass spectrometry" (multistage mass spectrometry) - allowed the researchers to collect information on the weights of the subunits of the rings as these units became shorter and shorter in the mass and increased in number with each additional cycle in the spectrometer. One of the computer scientists who joined the research team developed an algorithm that makes it possible to decipher the necessary reliable information from the mass of the spectrometer's results. With this method, the overlapping units were "glued" back together until they resembled a series of possible original ring structures, explains one of the researchers. The algorithm uses the information of the different unit masses of the tested proteins obtained at each step.
This work is a continuation of an award-winning automated approach that Bandeira, the principal investigator, and colleagues used to recover snake venom proteins. In their paper, the researchers explain how they used this new floor method to determine the structure of two different non-ribosomal proteins. In order to confirm their results, the researchers chose proteins that had already been deciphered in the past using a different, slow, tedious and expensive method using an NMR device. An NMR (Nuclear Magnetic Resonance) device allows obtaining information about the location of a particular atom within the compound while relying on the magnetic properties of the atom's nucleus. The team is currently working on more than ten additional proteins and has filed a patent application for this method.

2 תגובות
http://en.wikipedia.org/wiki/Nonribosomal_peptide
What is the difference between ribosomal proteins and non-ribosomal proteins? What does the "non-ribosomal nature of the proteins" mean?