How the protein factory combines amino acid molecules and creates proteins from them

Every kindergarten teacher knows how to teach kindergarten children how to string beads on a string and make a string. But in this way a random string is generated, or according to the preferences of the string rhymer. On the other hand, a protein molecule, built as a sort of string of "beads" which are amino acid molecules, is created in the ribosome, the cell's protein factory, according to an orderly plan dictated by the genetic code. But how exactly does the ribosome attach the amino acids to each other and create the protein "string"? How does he create the peptide bond between them? This question has been open for many years, and is still not completely clear, but recently some significant steps have been taken towards solving the mystery. Prof. Ada Yonat and the members of her research group from the Department of Structural Biology at the Weizmann Institute of Science, who worked in collaboration with Prof. Luis Massa from the State University of New York, and Jerome Carla, winner of the 1985 Nobel Prize in Physics from the US Naval Research Institutes, are responsible for this achievement.
In previous studies, Prof. Yonat was able to decipher the three-dimensional spatial structure of the ribosome, which is a structure containing several proteins, and clarify the principles of its operation.
Scientists from the Max Planck Institute in Germany also participated in this work, which was carried out in the past and lasted more than 20 years. In order to decipher the structure of the ribosome, the scientists used a technology called X-ray (X-ray) crystallography. In the first step of this method, crystals must be created from the protein, or from the studied particle. These crystals are irradiated with X-rays, and then the radiation scattered from the crystal is measured, the data is processed, and the density distribution of the measured material is mapped in space. This information teaches the scientists about the spatial structure of the molecules that make up the crystal.
This is how Prof. Yonat and the members of the research group she heads were able to decipher the structure of the ribosome, and then also "photograph" its subunits, called S30 and S50, in their active configurations. This is how the members of Prof. Yonat's research group were able to "see" the small subunit, at the stage when the first contact between the messenger RNA molecule and the ribosome was made. This contact signals the possibility of starting the process of protein production according to the genetic information carried in the messenger RNA molecule. Thus, in fact, the ribosome reads the molecular string of the messenger RNA, and according to the information contained in it, creates a protein molecular string, built from amino acids in a chemical process that is carried out in the large subunit of the ribosome.
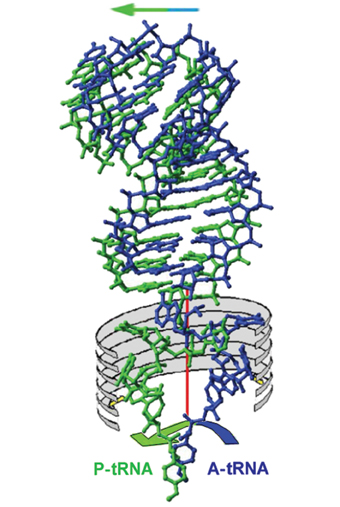
In the process of creating the protein in the ribosome, molecular "trucks" called tRNA are involved, which bring the amino acid molecules to the ribosome. In fact, each amino acid has a "truck" unique to it, which knows how to read only its coding, and only carries it. Despite this functional uniqueness, it turns out that basically, all the "trucks" have a similar structure. The scientists Aaron Kellogg, winner of the Nobel Prize in Chemistry in 1982, and Alexander Rich (both members of the Board of Directors of the Weizmann Institute of Science), who focused on the three-dimensional spatial structure of tRNA, discovered the structure of the "trucks" as they appear When not attached to the ribosome: they are mostly made up of a double strand of RNA, but the two ends with the unique properties and the chemical-biological function are made of a very flexible single-stranded chain.
The "truck" chosen according to the coding in the genetic code binds on the one hand to the messenger RNA molecule, and since it carries the appropriate amino acid, it actually leads the amino acid molecule to the nascent protein chain. When one such "truck" fulfills its role, it gives way to another "truck", which carries another amino acid, and so the process repeats until the completion of the protein chain.
As Kellogg and Rich discovered, the "truck" (TRNA) consists of two parts, one part whose structure can change relatively easily, while the other part has a more permanent structure. The permanent part includes a kind of "catch" that holds the amino acid molecule. Prof. Yonat and the members of her research group showed that, in the process of creating the peptide bond, one part of the tRNA moves forward together with the messenger RNA molecule (to advance the genetic code), while the other part (that is, the the single RNA) rotates, with the point of contact between the constant part and the single RNA strand serving as a pivot. Thus, at each turn, the rotating part of the "truck" brings another amino acid to the resulting protein. The rotational movement is possible because the two sites on the ribosome to which the moving unit of the tRNA binds are symmetrical, and in this way the direction of the rotational movement is dictated and supported by the framework which is the cornerstone of the ribosome structure.
The three-dimensional spatial structure of the free TRNA unit has been known for two decades. However, the changes that occur when binding to the ribosome are still being studied. The difficulty in deciphering the mystery stems from the fact that immediately upon its binding to messenger RNA (on the one hand) and to the amino acid (on the other) it begins to move and act very quickly. This speed makes it difficult for the scientists who try to "freeze" the unit in certain situations and decipher their structures. The members of Prof. Yonat's research group used molecules simulating it, and in this way were able to decipher the structure of the TRNA unit that carries the amino acid when it is bound to half of the active site of the "truck". After discovering the symmetry that causes the rotational motion, the scientists were able to calculate the position of the second TRNA unit. This discovery significantly advanced the research journey to unravel the mysteries of the peptide bond, but it was still unclear when and where the construction of the protein chain takes place. That is, whether the amino acids are attached to the emerging protein chain before, after, or during the rotation.
To answer this question, Prof. Yonat collaborated with the scientists Luis Massa from the State University of New York, and Jerome Carla, winner of the 1985 Nobel Prize in Physics from the US Naval Research Institutes. Together, they used a method called "quantum crystallography", based on the calculation of the quantum relationship between the atoms that make up the studied unit. In practice, they focused on the 50 atoms located around the region in the tRNA unit that makes the peptide bond, and calculated how these atoms would arrange themselves in space if they were given complete freedom. Under such conditions, the structure that the atoms will "prefer" is the structure whose maintenance will require the expenditure of the least amount of energy. This is how they managed to "catch" the TRNA in an intermediate state during the rotation. Another calculation showed that the energy needed for the process is produced, in addition to the energy "allocated" to drive the movement of the genetic code as a whole, also from the formation and breaking of chemical bonds between the rotating region and the frame of the ribosome during the rotation process. The findings of this study were recently published in the scientific journal "Records of the Academy of Sciences of the United States", PNAS.
 Prof. Yonat: "Certain antibiotic drugs harm bacteria when they stick to the binding site of the tRNA unit in their ribosomes, paralyzing them, thereby preventing the formation of new proteins essential to the bacteria. The understanding we achieved regarding the way protein is built in the ribosome will allow the development of more effective antibiotic drugs, which can also harm bacteria that have already developed resistance against existing antibiotic drugs."
Prof. Yonat: "Certain antibiotic drugs harm bacteria when they stick to the binding site of the tRNA unit in their ribosomes, paralyzing them, thereby preventing the formation of new proteins essential to the bacteria. The understanding we achieved regarding the way protein is built in the ribosome will allow the development of more effective antibiotic drugs, which can also harm bacteria that have already developed resistance against existing antibiotic drugs."
in the rotation process. The findings of this study were recently published in the scientific journal "Records of the Academy of Sciences of the United States", PNAS.

6 תגובות
I wouldn't relate to it but now I will relate to it much more.
Thank you!
You are XNUMX% right, I am also a kindergartener and I think so.