Weizmann Institute of Science scientists presented the possibility of reliable and accurate blood tests for cancer diagnosis
Blood tests for cancer diagnosis are the next big promise in medicine. Although these simple, accessible and non-invasive tests are already used in the early diagnosis of cancer, at this stage they are not reliable enough to enter widespread use. A new method developed by Weizmann Institute of Science scientists may lead to a leap forward in the field and the development of blood tests to diagnose cancer with an unprecedented level of accuracy. The research findings are published today in the scientific journal Nature Biotechnology.
"The currently accepted methods for detecting cancer are often invasive and unpleasant or alternatively expensive and inaccessible," says the head of the research team, Dr. Efrat heard from the department of immunology and biological regeneration of the institute. For example, taking a tissue sample (biopsy) - whether surgically or using a long needle - can be painful and sometimes even involves a certain risk. In contrast, imaging tests such as MRI or PET-CT rely on expensive and cumbersome equipment that is not available everywhere. Therefore, effective blood tests, also known as liquid biopsy, may be a successful alternative to all of these. "It's not just about convenience. Reducing the discomfort associated with the test greatly increases the chances that people will want to go get tested, and accordingly also increases the chance of early detection and saving lives," adds Dr. Shema.
The idea of diagnosing cancer through blood tests is based on the fact that the byproducts of cell destruction are floating in our blood fluid: DNA segments and proteins derived from dead blood cells. When cancer develops in the body, fragments of DNA and proteins originating from dead tumor cells also float in the blood fluid. Various blood tests for cancer diagnosis are currently in advanced stages of development, but most of them suffer from significant shortcomings. The first generation of these tests relied on the identification of genetic mutations that are very difficult to detect in blood tests, since they constitute only a fraction of the genetic material secreted into the blood; Moreover, the mere existence of the mutations does not necessarily indicate the development of cancer. The new generation of blood tests focuses on a different type of data: epigenetic mechanisms, meaning changes in the cell genome that do not involve changes in the DNA itself, for example, various chemical tags that stick to the DNA molecule and thus affect gene expression. But even these methods have notable drawbacks: some tests require a large amount of blood, while others rely on the detection of only one type of epigenetic change, so their results are not very reliable.
In the current study, conducted under the leadership of research students Vadim Padiuk and Nir Erez, the researchers aimed to develop a new approach that would make it possible to reliably diagnose cancer based on a small amount of blood. To this end, they relied on a method for imaging single molecules, which Dr. Shema developed during her post-doctoral research period at Harvard Medical School and the Broad Institute in Boston. This method, which is aided by fluorescence microscopy, makes it possible to carry out accurate and precise epigenetic mapping on the basis of a very small sample. Dr. Shema hypothesized that this method could be applied to examine epigenetic changes in nucleosomes - the basic subunits that make up the packaging of the genetic material in the cell. Like floating remains from a sunken ship, millions of nucleosomes of dead cells are released into the bloodstream and may be fertile ground for cancer diagnosis.
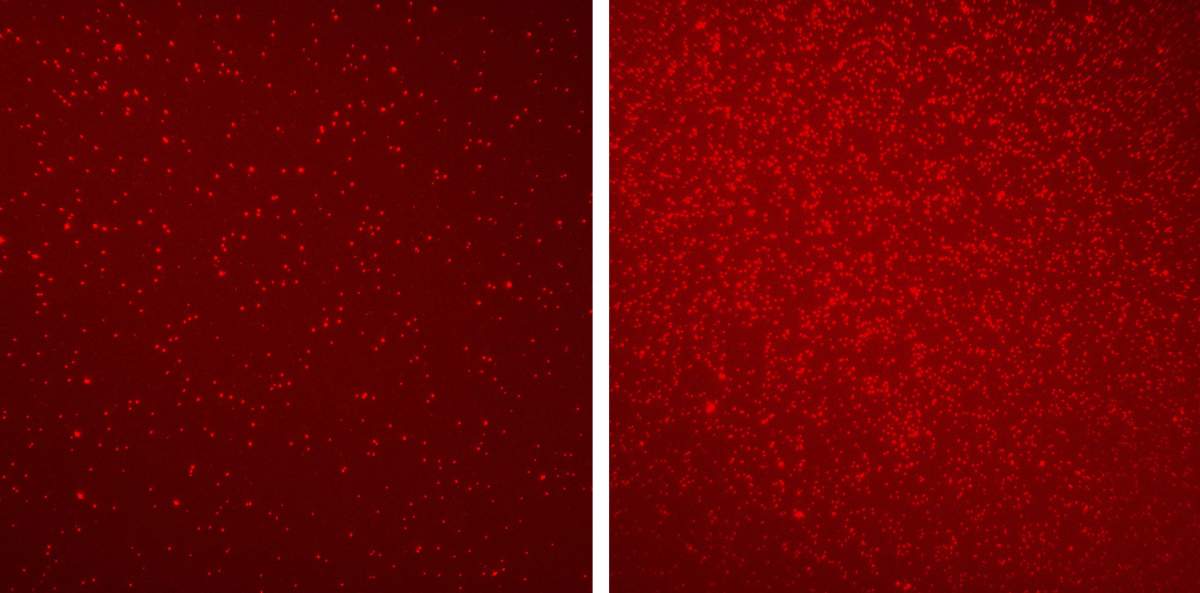
In the current study, Padiuk and Erez, along with their colleagues, used Dr. Shema's method to compare the nucleosomes from the blood of 30 healthy volunteers with those of 60 colon cancer patients at different stages of the disease. The scientists identified a fundamentally different epigenetic pattern in each of the groups. Using a small amount of blood, they were able to map six epigenetic changes associated with cancer and even identify a variety of other known cancer markers in the samples, including protein fragments from dead tumors that cannot be detected with conventional technologies.
Using artificial intelligence algorithms, the scientists, in collaboration with Prof. Guy Ron from the Rakah Institute of Physics of the Hebrew University of Jerusalem, analyzed the massive database received from the two groups, and even tested different combinations of epigenetic changes. At the same time, to make sure that the findings are relevant not only to colon tumors, they made comparisons between the nucleosomes from the blood of healthy people and those from the blood of ten pancreatic cancer patients. "Our algorithm knew how to distinguish between groups of healthy people and cancer patients with an accuracy of 92 percent - an unprecedented level of confidence for this type of testing," says Dr. Shema.
If these findings are reproduced in larger experiments, the way will be paved for the development of a multi-parameter blood test for cancer detection based on a blood sample of less than one milliliter. Tests of this type are expected to advance the world of personalized medicine as well, since the multitude of data revealed in the test is expected to allow optimal treatment to be tailored to each patient.
"We have presented proof of the feasibility of our method, and now it remains to be seen whether the findings will also be reproduced in the clinical trials," concludes Dr. Shema. "In the future, this method may make it possible to diagnose not only different types of cancer, but also other diseases that leave traces in the blood, such as autoimmune diseases and heart diseases."
Dr. Noa Pirat and Olga Barash from the Department of Immunology and Biological Regeneration also participated in the study; Dr. Ekaterina Andriaisheva, Abhijit Shinde and Daniel Jones of SeqLL Inc. in Woburn, Massachusetts; Dr. Barak Zakai and Dr. Yael Mabor from Kaplan Medical Center; and Dr. Tamar Peretz, Prof. Ila Hubert, Dr. Jonathan Cohen, Dr. Azam Salah, Dr. Mark Temper, Dr. Albert Grinsfon, Miriam Maoz and Dr. Aviad Zik from the Hadassah University Medical Center.
It should be noted that this week Tel Aviv University also issued an announcement about research in the same field: A simple blood test to detect cancer instead of invasive tests
More of the topic in Hayadan:
