Researchers from the University of Washington seek to harness the skills and motivation of those gamers in order to reach new medical discoveries in reality
Persistent computer players (gamers) have spent many years of collective brainpower trying to rescue princesses in distress and protect the Earth from alien invasion. All this, in front of the computer screen. But now researchers from the University of Washington are trying to harness the skills and motivation of those gamers in order to reach new medical discoveries in reality.
A new game called Foldit turns the complicated act of folding proteins into a competitive sport. The first steps guide the players and teach them the laws, identical to the laws of physics and chemistry, which cause the protein strands to curl and curl into incredibly intricate three-dimensional shapes.
There are millions of possibilities for the folding of any protein, but usually it can perform its action in the body only when it is folded in exactly the right way. Although we know the DNA sequences of many proteins, we still do not know how they fold into the complex shapes that allow them to perform their action in cells and the body. Since most of the operations in human cells are carried out by the proteins, understanding the way they fold and organize can jump medical science forward in many years.
"We hope to change the way science is done, and by whom it is done," says Zoran Popovic, an associate professor at the University of Washington of Engineering and Computer Science and one of the authors of the paper. "Our ultimate goal is to get regular people playing the game, and eventually being nominated for a Nobel Prize."
Computer simulations are currently trying to process all possible forms of proteins, but this mathematical problem is so complicated that even if all the computers in the world worked on it together, it would still take hundreds of years to solve it. In 2005, David Baker, a professor of biochemistry at the University of Washington, developed a project called Rosetta@home, which uses the computing power of computers around the world to solve the protein folding problem. But even the 200,000 dedicated volunteers who donated their computing time to the cause are not enough to solve this problem.
"There are too many options for the computer to go through every single one," says Baker. "The Rosetta@home approach works well on small proteins, but as the protein gets bigger and bigger it gets harder and the computers often fail."
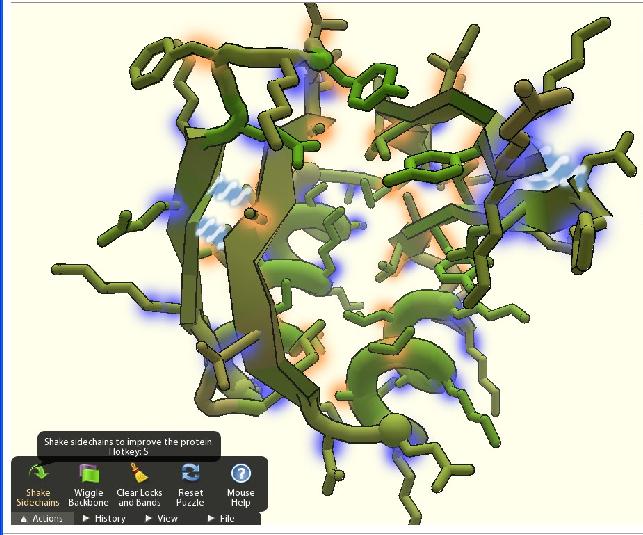
The Rosetta Project used to display the forms of proteins that the computer was working on as a screen saver. Baker says that he has received e-mails from people who said that they can understand how the protein should fold, but are frustrated by their inability to transfer the instructions to the software. The idea for the Foldit game was born, among other things, from those people.
The game uses the same protein folding software used by the Rosetta Project, but it allows players to manipulate the proteins displayed on the screen themselves. The game takes advantage of the natural ability of humans to solve problems in a XNUMXD environment - a feature that is very difficult to impart to computers.
"People who use their intuition may come up with the right answer in a much shorter time," says Baker. The intuitive skills needed from a good Foldit player are not necessarily the same as those needed from biologists or scientists. Baker says his 13-year-old son is faster at folding proteins than he is. Others may be even faster.
"I imagine that there is a 12-year-old boy in Indonesia who can see all this in his head," says Baker. The researchers hope to advance science by discovering extraordinary talents in protein folding, who are naturally able to see proteins in three dimensions. "Some people are able to just look at a game, and in less than two minutes get the highest score," says Popovich. "They can't even explain what they're doing, but somehow they're able to do it."
A look at the game itself reveals a particularly complicated version of XNUMXD Tetris. Sheets, ropes and geometric shapes of different colors crowd the screen, and the player must move them in the right way in order to advance to the next level. The current stages bring as a challenge proteins with a known structure, but starting this week the developers will allow the general public free access to the game, and will offer proteins whose structures are unknown to science. In addition, starting this week, teams of Foldit players will compete against other research teams around the world, in the major protein structure championship, which will be held every two years.
In a few months the developers plan to add new challenges to Foldit. Among other things, they intend to offer the possibility of creating new proteins that we would like to have. These new enzymes could break down industrial waste, for example, or absorb carbon dioxide from the air. Although Baker recently succeeded in creating two new enzymes, in collaboration with the group of Dan Tofik from the Weizmann Institute and Kendall Hawke from the University of California, Los Angeles, the planning of these enzymes was done with the help of extremely powerful supercomputers and required a lot of time. The game developers hope that human players will be able to solve the problem quickly and efficiently.
Ultimately, the researchers hope that they can take an existing medical problem, such as the HIV virus, and present it as a challenge for the players. The players from all over the world will have to design a protein with a special shape that can attach to the proteins of the virus and paralyze them. The most successful charts will be synthesized in Baker's lab and tested in growth plates. Players with extremely high scores will be recognized in scientific articles that will most likely be published in the world's most important scientific journals. More importantly, they will get to participate in the global scientific effort to promote health and science - and all this without even requiring a bachelor's degree.
The game can be downloaded for free on the website fold.it.
Demo video of the game
Learn how to play an expert protein folding game

18 תגובות
refresh,
There are achievements and also quite significant ones you can check on their website.
Roy,
There is no need to sacrifice research grants. Surely pharmaceutical companies will be happy to pay for marketable results.
I wonder if there are already significant achievements for gamers who play this? A follow-up is needed here.
Wow, it looks cool to God, especially since my lab is dealing with exactly this topic. From today I can say that we are simply playing and trying to compete with 13-year-old children..
"Fate has a developed sense of irony" - who would have believed that gamers who waste their time in an imaginary world could bring real benefit to humanity.
Cool,
I know some things in biology, but even I had a hard time understanding how things work exactly in the game. I guess with enough free time and play, you eventually start to see the necessary patterns.
-------
Science for everyone: http://www.israelscience.weebly.com
Okay, I don't understand anything about biology, but let's face it, there's really nothing to understand, just go through the training steps and then you'll get the idea - that everything should be straight and tight as possible
I've only played a few hours in Kollel and I'm 400th place so it's not that bad even without a background
I played and I didn't understand what they wanted from me.. there is a situation that it is because I have no background in biology.
There are two types of branches and in any case do all the branches need to be far from each other?
And some hydrogen bonds suddenly appear.
And if possible, a brief explanation?
I played. Definitely a fun game. I even recommended it to my students.
-------
Science for everyone: http://www.israelscience.weebly.com
Someone got to play it, it's one of the original things!! 1
Believe me I thought about this idea (and even recommended it to some professor who never responded to the email I sent him).
On the one hand, it's a shame.
On the other hand - what a beauty that it was implemented 🙂
Are you sure it's not April 1st?
exciting. In this business money will be and will be.
Ami,
You can't disagree with my opinion, because I agree with your opinion. Money is one of the most attractive factors that can be offered. At the same time, I don't think Baker's lab has the will or the ability to offer the sums of money from the grants they have. It will have to become something more industrial.
-------
Science for everyone: http://www.israelscience.weebly.com
There is no doubt that I will try this software, if I can contribute at such significant levels.
I really want to start, wish me luck 🙂
Roy, I disagree with you: money is the cheapest item in this loop. It is available and accessible and saves even more money when invested correctly. Reputation is great, but what will the hungry 2.0D geniuses of the third world do with reputation? Such a style XNUMX protein folding project could, potentially, yield results that would in turn yield more public interest (or in other words = more money). I believe that the cheapest way, in this case would be to distribute as much money as possible to as many people as possible as a function of their (actual) contribution to research development.
With the blessing "Capitalism is not a wall and I am an air hammer"
Ami Bachar
Money is expensive. Reputation comes for free. Baker won't offer budgets from his lab to anyone who plays.
I believe that when we reach the stage where there will be a global competition of the game, the organizers will find additional funding sources that will give especially large financial prizes to the outstanding players.
I agree with Ami. A wonderful idea, no doubt.
Amazing. It could have a great future if the reward was a little more practical than an essay or the imaginary Nobel Prize option. Of money. Money must be given for successes. Or then we will discover all those folding geniuses with supreme grace who are able to give a quick answer to what no computer can give, and all this after a day of work in the field under the hot sun.