Scientists "calculated backwards" the molecular history of a certain protein, discovered the roots of its structure, and succeeded in reproducing in the laboratory the evolutionary process that led to the actual creation of the modern protein.
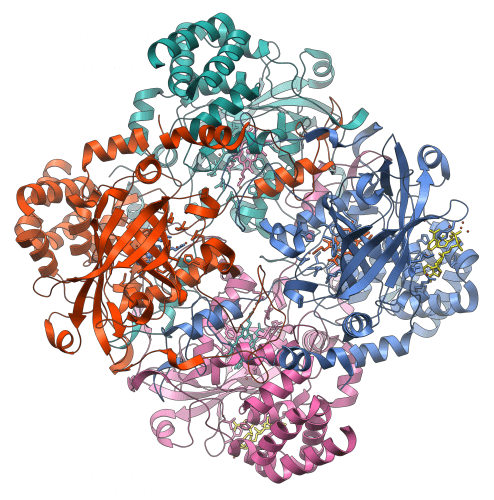
How was a song born? like the laugh. It starts from the inside, and rolls out. And how is protein born? like a propeller It starts small, and multiplies throughout. This is a possibility that emerges from original research by Prof. Dan Tofik, from the Department of Biomolecular Sciences at the Weizmann Institute of Science. Proteins are the molecular machines that drive the life processes in the cells of plants, animals and humans. Certain changes in the structure of the protein, or in the sequence of the amino acids that make it up, may cause disorders, diseases, and even death. On the other hand, changes in the protein structures are also the growth engine of evolution - they are the ones that create new opportunities that are brought before the court of natural selection.
The question of the formation of the proteins is, therefore, a key question in understanding the processes of life. So how is protein really born? It is common to assume that every protein has a "father": another protein, older than it, in which a mutation or an error in replication occurred in the gene encoding it, which caused the formation of the new protein. This is how things went generation after generation, until the proteins we know today were created. But how is a protein "ish ma'in" created? And how were the earlier proteins created, i.e. the first proteins in the evolutionary chain, those ancestors from which the later proteins developed? In order to answer this question, Prof. Toufik and the members of the research group he heads set out on a kind of time journey, which begins with a calculation - and ends with an experiment.
An intact protein is a complex and large structure, and the likelihood that it is formed by spontaneous self-assembly is somewhat similar to the possibility that a large airliner will spontaneously assemble from a scrap pile after being hit by a tornado. "But", says Prof. Toufik, "it is known that peptides, which are a kind of relatively short protein sequences that do not have a defined spatial structure (that is, they do not have a functional fold), are formed from time to time, under certain conditions, in nature and in the laboratory." Hence the hypothesis arose, that such peptide sequences may adhere to each other, and thus, as a result of the fusion, arrive together at the structure of a protein molecule. At this stage, the various weak forces that act between the peptides will cause them to fold, and create the spatial structure, which under certain conditions may have a certain function.
The scientists chose to examine the origins of the development of a propeller-like protein, built from several subunits. They used a calculation method invented by Linus Pauling, which allows to calculate, step by step, the state or the molecular configuration that preceded the sequences given today. This way it is possible to "go back in time", and discover the roots of molecular structures - for example, proteins from a certain family - that we know today. In this way, they were able to identify the peptide "blade", which is made up of only about 50 amino acids, which is the ancient root of a certain propeller protein, which could have been created through the fusion and self-assembly of peptides - or "blades" - of this type.
At this point, the scientists reversed the direction of movement of their "time machine". That is, instead of calculating the evolution processes of the peptide (the "blade") backwards, they moved to a laboratory experiment, in which they tested whether a certain peptide could actually merge with other peptides, and create a protein together.
They started with a peptide (blade) that was about 50 amino acids long, and let it evolve in vitro. This is a method in which the scientists create different molecular sequences, which are exposed to mutations and undergo "improvement" generation after generation. The method allows, in fact, to have a kind of accelerated evolution. This is how they managed to reproduce the process that apparently took place in nature: a peptide ("blade") of about 50 amino acids underwent spontaneous assembly, until the formation of a protein with a contemporary structure, consisting of about 250 amino acids. Tracking the process has also allowed scientists to understand what forces shape the process of creating an intact primary protein.
Later, the members of the research group examined bacterial genomes, and discovered in them clear evidence of the existence of an evolutionary process very similar to the one they had in the laboratory experiment of duplication and conjugation of short molecular sequences.
* The quote that opens this article is taken from the song "Ich Shir Nold" by Yonatan Gefen, and is included in the show "The Sixteenth Lamb".
Science books
3 significant developmental steps were required to get from the initial peptide to the functional modern protein. The accelerated evolution lasted about 5 months, while in nature it probably took several million years.
