A new study reveals that among the population of bacteria there are those that survive even after exposure to antibiotic treatment, because of their slower development. The finding may help in the fight against the increasing resistance of bacteria
Merit Sloin
In recent years, the problem of antibiotic resistance is getting worse. The predatory bacteria claim victims, premature babies are infected with violent bacteria, and science often stands helpless in the face of bacteria that are resistant to all antibiotic drugs. How do the tiny creatures manage to outwit the scientists and escape the antibiotics? The answer is known: within a population of bacteria exposed to antibiotics there are those that have undergone a mutation, which gives them resistance to antibiotics. Therefore, in the presence of the antibiotic substance, they will survive and multiply, while their friends who did not undergo the mutation will become extinct. This is how a bacterial population is obtained, most of which consists of resistant bacteria. The trait of resistance is passed on to the offspring and a new strain of bacteria is created, resistant to antibiotics.
From this description it is possible to understand that as long as no mutation occurred the bacteria will all become extinct. But it turns out that this is not necessarily so. In a study conducted by an Israeli researcher and a team of researchers from the Rockefeller University in the USA, and which was recently published in "Science", it was found that in every bacterial population there are few bacteria that survive exposure to antibiotic treatment without mutating. Their tiny number allows them not to be detected in the tests done for the development of new antibiotics.
"The problem of bacterial resistance to antibiotics is becoming more complex every day," explains Dr. Nathalie Laban from the Rakeh Institute of Physics at the Hebrew University, who conducted the research. "Resistance is created quickly. Therefore, for example, there is a position according to which, since most antibiotic substances come from nature, the genes for resistance already exist in bacteria." Based on this position, pharmaceutical companies decided to develop synthetic antibiotic substances. About two years ago, such a new substance called "linezolid" was put into use, high hopes were planted in it, but in less than two years bacteria resistant to it developed. This is an example of the difficulty involved in the development of new antibiotics: the cost of its development is enormous, the uncertainty is great, and so it happens that, paradoxically, the pharmaceutical companies reduce their investments in the development of new antibiotics at the same time as the resistance of the bacteria increases.
"One of the main tasks is to reach a deep understanding of how antibiotic resistance develops. This is a critical step before a new antibiotic is introduced to the market," says Balban. "The goal is to get rid of the entire population of bacteria, so we wanted to understand why some bacteria react differently to antibiotic treatment. When we looked in our study at the dynamics of the bacterial population, we saw that most of the population dies very quickly, but there are also bacteria in it that the antibiotics don't work on. These are 'slow' bacteria - they develop slowly and therefore are not harmed by the antibiotics, which work on rapidly dividing cells. The developers of the antibiotics did not know until now that these bacteria had to be taken into account, even though these findings were hidden between the lines of the medical literature in articles published at the beginning of the last century."
Antibiotics were introduced in World War II. Army doctor Joseph Bigger noticed that after applying antibiotics to the wounds of war wounded, the wound froze. But after a short time he reappeared. When he isolated the bacteria from the wound and treated them with antibiotics, he expected to find resistance of the entire population to antibiotics, because he knew that those that survived after the treatment were actually those bacteria that had undergone a mutation that gave them resistance. But Bigger found out that not all the bacteria that survived were resistant, and in fact he found a mixed population of resistant and non-resistant bacteria. He did not know how to explain the find, but reported it in the scientific literature.
This phenomenon has not received special interest from scientists. Only in the last ten years have they become interested in the non-uniform response of bacteria to antibiotics. Five years ago, Laban entered the field. "Our basic approach was to try to describe the phenomenon of resistance using mathematical tools, in order to reach a deeper understanding of it," she says. Together with the team of researchers from the USA, she built a mathematical model that describes the behavior of bacterial populations.
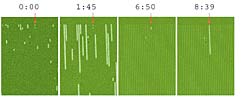
"The model hinted that in every bacterial population there are subpopulations that do not grow at the same rate and they are the ones that survive the antibiotics," says Laban. "We decided to check the phenomenon in a quantitative and precise way." To this end, the researchers developed a chip with tiny channels, in each of which a single bacterium grows. Using a microscope it is possible to trace the behavior of each of the bacteria and see how it reacts to antibiotics. "We saw that within the group there were some bacteria, which after the administration of antibiotics were the only ones that survived. When we looked back in time, we saw that they were there even before the antibiotics were given, but developed slowly compared to their friends," says Balban. "The slow bacteria found in the population are therefore the ones that survive the antibiotic treatment. We know that antibiotics mainly damage rapidly dividing cells. This is the reason, then, that the slow cells are not damaged, and after antibiotic treatment they survive and the disease returns. The slow-moving bacteria are found in every bacterial population, regardless of whether an antibiotic is given or not, and we speculate that these are the bacteria that may accumulate mutations and acquire resistance to antibiotics."
The findings of the group of researchers also have practical significance. "A different approach is needed for the development of antibiotic drugs or antibiotic companion drugs," she says. "Instead of referring only to the damage they cause to the majority of the bacterial population, one should also refer to the damage they cause to the slow bacteria, with the aim of preventing them from surviving after treatment. This can be done in two ways: develop antibiotics that also kill the slow bacteria, or find a way to increase their rate of development and then attack them with regular antibiotics."
The chips on which the bacteria develop can, by the way, be used for another purpose: rapid identification of infectious diseases. The fact that they provide immediate information on the sensitivity of bacteria to antibiotics, before the bacteria multiply into visible colonies, could make it possible to save the two or three days currently required for such a diagnosis.
Behavior of bacteria in response to antibiotic treatment. The chip in sequence
The images contain rows, in each of which a single bacterium develops
starting position
The bacteria develop on the chip; A slow bacterium (marked by an arrow) lags behind the rest
after administration of antibiotics; All the bacteria die, except for the slow bacteria
The bacterium that survived continues to develop after the antibiotic is removed
https://www.hayadan.org.il/BuildaGate4/general2/data_card.php?Cat=~~~145223961~~~152&SiteName=hayadan
