Weizmann Institute of Science scientists have developed a living cell-like system, on a chip, in which the small unit of a bacterial ribosome is created and assembled - autonomously
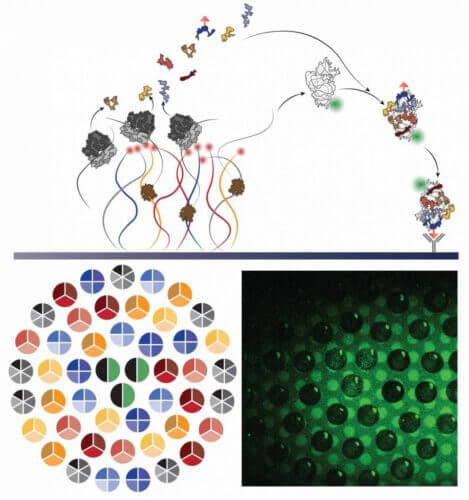
The ribosome, the cell's protein factory, is the only machine in nature that produces its own parts. How does she do it? The answer to this question may shed light on the original processes of the development of life, as well as lead to the development of advanced ways to design medicines and more. The research group of Prof. Roi Bar-Ziv and staff scientist Dr. Shirley Shulman Dauba, in the Department of Chemical and Biological Physics at the Weizmann Institute of Science, have been facing this challenge for many years. Recently, after a research effort that lasted about seven years, the scientists succeeded in building a living cell-like system, which operates on a chip, and in which the small unit of the ribosome called s30 is created and assembled autonomously.
The s30 unit is made up of a long strand of the nucleic acid RNA, to which 20 different proteins are bound, in a defined and precise structure. The weak forces (attraction and repulsion) acting between the different protein molecules and the RNA strand influence and cause the proteins and RNA to organize together in the correct structure, which is essential for the proper functioning of the ribosome. Also involved in the process are six "companion proteins" ("chaperones"), which help the proteins and RNA to organize themselves, thus speeding up the assembly process. That is, in the formation of the s30 unit, at least 27 genes are involved that encode all the components of the unit. The biggest challenge in recreating an autonomous process for the production and construction of the ribosome, has so far been to find optimal conditions for the production of RNA and the many proteins in a way that would allow efficient assembly of the complete unit.
Prof. Bar-Ziv and Dr. Shulman Dauba, the post-doctoral researcher Dr. Michael Levy (who led the research) and the research student Reuven Falkovitz created an artificial cellular system, capable of imitating the natural process of production and assembly of ribosomes and thus overcoming the challenge of efficiency and quantities . The system is based on a tiny chip, on which dense DNA strands are fixed. In this study, the genes incorporated into the device are the same 27 genes, which carry the information needed to produce the components of the small ribosome unit of the E. coli bacterium. To allow the genes to "express" the information encoded in them and to create the proteins and the RNA strand according to it, the scientists flowed into the chip a solution containing the entire mechanism of E. coli bacteria, which is responsible for translating proteins (including ribosomes). The newly formed ribosomal units were captured on a surface of "molecular traps" in close proximity to their genes, which improved the overall efficiency of the assembly, and enabled real-time observation of the process. At this point, the scientists "take a step back", and allow the tiny biological device, which simulates a living cell, to operate autonomously and create the proteins and the RNA strand without any intervention on their part.
Molecular hierarchy
However, being able to produce all the components of the s30 unit is just the beginning. How can these components be made to fit together in the right way, which will indeed create the correct and active structure of the unit? Prof. Bar-Ziv, Dr. Shulman Dauba and the members of their research group at the institute worked on solving this mystery for seven years.
During hundreds of experiments, the scientists were able to decipher, tap by tap, the correct pattern in which the various genes should be placed on the chip. The organization of the genes on the chip, similar to the organization of genes on chromosomes, affects the timing of the formation of the proteins and their relative location and quantity. Using the fixation of different clusters of genes in the same device, each cluster with a different composition of genes, the scientists revealed the construction process of the ribosome unit. That is, by recreating the defined and hierarchical process of assembling the s30 unit, step by step, it was possible to prove that the unit was assembled. With this method it is also possible to reveal the effect of the auxiliary proteins on the process.
Prof. Bar-Ziv and Dr. Shulman Dauba say that this discovery may enable the production and assembly of various multi-molecular structures, natural and even those that do not exist in nature. Among other things, it will be possible to create structures of disease-causing bacteria or viruses in this way, without risking the morbidity that the entire bacteria or viruses may cause. This is because only synthetic genes are needed to initiate production and assembly. This ability may lead, in the future, to the development of medicines and the advancement of production lines for various materials and molecular components necessary for industry.
More of the topic in Hayadan:
