Only 1% of the human genome contains genes that code for proteins and almost half of the genome contains genes used for control. More than half of the human genome consists of repetitive sequences that could not be sequenced in the first pass over the human genome in the nineties, but now technology makes this possible and a new era of genome research has opened
By: Gabriel Hartley, PhD Student in Molecular and Cellular Biology, University of Connecticut. Translation: Avi Blizovsky, The Knowledge Site
When the heads of the Human Genome Project announced in 2003 that they had completed the sequencing of the first human genome, it was an important achievement: for the first time, the encryption of the DNA software on which human life is based was deciphered. But the announcement came with a catch - they were unable to assemble all the genetic information stored in the genome. There were gaps: it was often repetitive areas that were confusing to put them together and understand where they belonged.
With the advancement of technology that now makes it possible to deal with repetitive sequences, scientists finally filled in these gaps in May 2021, and the first end-to-end human genome was officially published on March 31, 2022.
I am a genome biologist who studies repetitive DNA sequences and how they shape genomes throughout evolutionary history. I was part of the team that helped characterize the repetitive sequences in the genome. And now that we have a truly complete human genome, the repetitive regions that have finally been revealed are being fully explored for the first time.
The missing pieces of the puzzle
The German botanist Hans Winkler coined the word "genome" in 1920, when he combined the word "gene" with the suffix "-ome", which means "complete set", to describe the complete DNA sequence found in each cell. Researchers still use this word a century later to refer to the genetic material that makes up an organism.
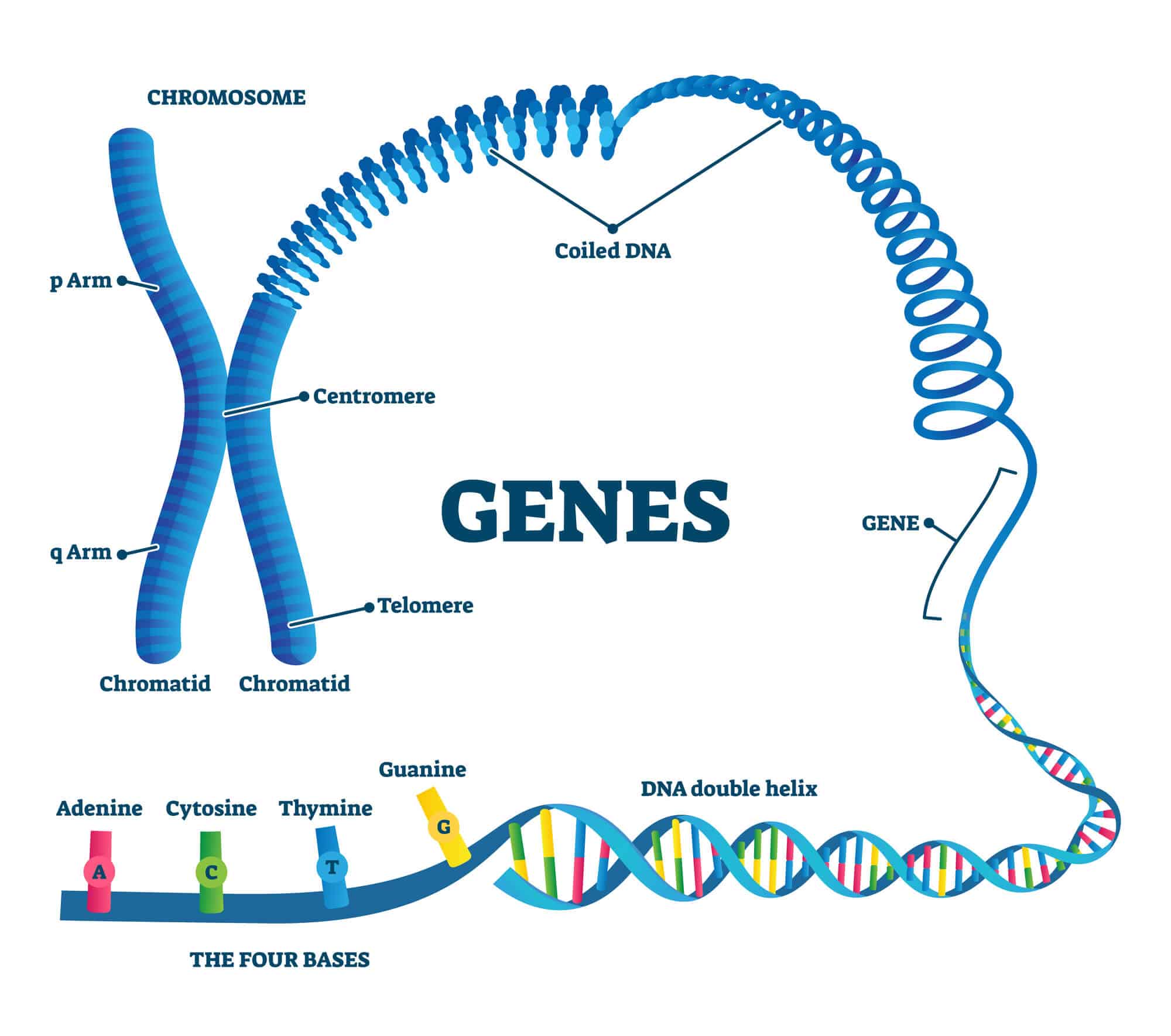
One way to describe what a genome looks like is to compare it to a reference book. In this analogy, a genome is an anthology containing the DNA instructions for life. It consists of a huge variety of nucleotides (letters) that are packed into chromosomes (chapters). Each chromosome contains genes (paragraphs) which are regions of DNA that code for the specific proteins that allow the organism to function.
Diagram of a chromosome unraveled into coiled DNA, genes and nucleotides
Every living organism has a genome, but the size of the genome varies from species to species. An elephant uses the same form of genetic information as the grass it eats and the bacteria in its gut. But no two genomes look exactly the same. Some of them are short, like the genome of the bacteria of the species
residing within insects containing only 137 genes over 112,000 nucleotides. Some, like the 149 billion nucleotides of the flowering plant Paris japonica, are so long that it is hard to tell how many genes they contain.
But genes as they have been traditionally understood - segments of DNA that code for proteins - are only a small part of an organism's genome. In fact, they make up less than 2% of human DNA.
The human genome contains approximately 3 billion nucleotides and just under 20,000 protein-coding genes that account for approximately 1% of the total length of the genome. The remaining 99% are DNA sequences that do not code for proteins. Some of them are control elements that control how other genes work. Others are pseudogenes or genomic remnants that have lost their ability to function. More than half of the human genome consists of repetitive genes, with multiple copies of nearly identical sequences.
What are repetitive DNA segments?
The simplest form of repetitive DNA are blocks of DNA that repeat themselves over and over again in the same order and are called satellites. The number of satellite DNA segments varies from person to person but they often cluster near the ends of chromosomes in regions called telomeres. These regions protect the chromosomes from disruption during DNA replication. They are also found at the centromeres of chromosomes, a region that helps keep genetic information intact when cells divide.
Researchers still do not clearly understand all the functions of satellite DNA, but because satellite DNA creates unique patterns in each person, forensic biologists and genealogists use this genomic "fingerprint" to match crime scene samples to suspects and trace their origins. More than 50 genetic disorders are associated with changes in satellite DNA, including Huntington's disease.
Another common type of repetitive DNA contains interchangeable elements or sequences that can move along a genome. Some scientists have described them as selfish DNA because they can insert themselves anywhere in the genome, regardless of the consequences of doing so. As the human genome has evolved, many displaceable sequences have accumulated mutations that suppress their ability to move to avoid harmful interference, but it is likely that some can still move. For example, genes that can be replaced or shifted are associated with several diseases such as hemophilia A or genetic bleeding disorders.
Replaceable DNA is perhaps the reason why humans have a coccyx but no tail?
But interchangeable genomic elements aren't just disruptive. They can have regulatory functions that help control the expression of other DNA sequences. When concentrated at the centromeres, they may also help maintain the integrity of basic genes and contribute to cell survival.
The activity of replaceable genes can also contribute to evolution. Researchers have recently discovered that the insertion of a transposable element into a gene important to development may be the reason why some primates, including humans, no longer have tails. Rearrangement of chromosomes due to the introgression of exchangeable elements is even linked to the formation of new species such as the gibbons of Southeast Asia and the wallaby of Australia.
Completing the genomic puzzle
Until recently, many of these complex regions could be compared to the far side of the Moon: known to exist but not understood with great precision.
When the Human Genome Project was first launched in 1990, technological limitations made it impossible to fully uncover repetitive regions in the genome. Available sequencing technology could only read about 500 nucleotides at a time, and these short segments had to overlap each other. The researchers used these overlapping segments to identify the next nucleotides in the sequence, gradually expanding the assembly of the genome segment by segment.
Assembling the repetitive regions of the genome was like putting together a 1,000-piece jigsaw puzzle in which a cloudy sky is drawn: when every piece looks the same, how do you know where one cloud begins and another ends? With nearly identical overlapping parts in many places, complete sequencing of the genome by the first genome sequencers became impossible. Millions of nucleotides remained hidden in the first iteration of decoding the human genome.
Since then, the gaps in the human genome have been gradually filled. In 2021, the Telomere to Telomere Consortium (T2T), an international consortium of scientists working to complete the end-to-end assembly of the human genome, announced that all remaining gaps had finally been filled.
With the completion of the first human genome, researchers are now looking to understand the full diversity of humanity.
This has been made possible by improved sequencing technology capable of reading longer sequences of thousands of nucleotides along with additional information to place repetitive sequences within a larger picture where it is easier to identify their correct place in the genome. It's like turning a 1,000 piece puzzle into a 100 piece puzzle. The ability to read long sequences made it possible for the first time to assemble large repeating regions.
With the increasing power of long-term DNA sequencing technology, a new era has opened in genomics research - the possibility of deciphering for the first time complex sequences of repetitive genes across populations and species, and in a complete human genome without gaps. This knowledge provides valuable power for researchers to study repetitive regions and thereby create variations in genetic structure, making it possible to study species evolution and contribute to human health.
But one complete genome does not give the whole picture. Efforts are constantly being made to create diverse genomic reference sequences that fully represent the human population and life on Earth. Thanks to more complete genomic references obtained by the "telomere to telomere" method, scientists' understanding of the "dark matter" of the repetitive genome will become clearer.
For an article in The Conversation
More of the topic in Hayadan:
- The mapping of the human genome has been completed, humans are identical in 99.9 percent of their genes (2001)
- How was the first DNA molecule created?
- Molecular evolution eliminates lethal sequences
- Clone mammoths to protect the climate?
- Researchers have developed an algorithm that reads our DNA as human language
- The next phase of the human genome project was launched: haplotype mapping

2 תגובות
A beautiful and clear article at eye level.
What is its size in megabytes?