The researchers conclude and say: "Future research using high-speed atomic force microscopy (HS-AFM) will provide additional important information for understanding the dynamic activity of these complex translation machines"
[Translation by Dr. Moshe Nachmani]
An innovative technology of extremely fast atomic force microscopy allows to see clearly and visually the actions that take place in the cellular factory called the ribosome
Ribosomes are complex structures of proteins and nucleic acids that function as the main "factory" for protein synthesis inside the cells. At the same time, in the absence of conclusive evidence, the mode of activity of these structures is still subject to scientific disagreements. Now, researchers from Japan have been able to observe the structural dynamics that occur in ribosomes while building the proteins inside them.
Ribosomes were first discovered in the fifties of the last century and their extensive function was correctly understood for a long time - they "read" messenger RNA sequences (a type of RNA) and accordingly use specially ordered sequences of amino acids to produce new proteins. In particular, the main protein in the ribosome plays an integral role in the protein synthesis process by using protein factors responsible for the translation and extension of the amino acid sequence. However, until now it has been extremely difficult and challenging to decipher the exact structure of this major protein due to its high complexity. At this point, the high resolution and rapid image generation of an ultrafast atomic force microscope provides great value.
Atomic force microscopy uses a nanometer tip to "feel" the samples being tested, similar to the needle of vinyl record players that scans the record and receives sound information from it, except for the difference that the details revealed by an atomic force microscope can be at the level of atomic separation. The versatility of the method for different substrates already provided a great advantage for a variety of biological studies, but thanks to the improvement of the scanning speed, the method was able to capture dynamic processes, for the first time ever. The researchers were able to reveal with the help of the innovative method that the main protein actually jumps between two configurations (conformations) - one that corresponds to the previous structural models and a second that is new and completely unexpected. As for how the ribosome works - in the past a two-step mechanism was proposed to describe how the genetic information is translated by proteins called translational GTPase factors.
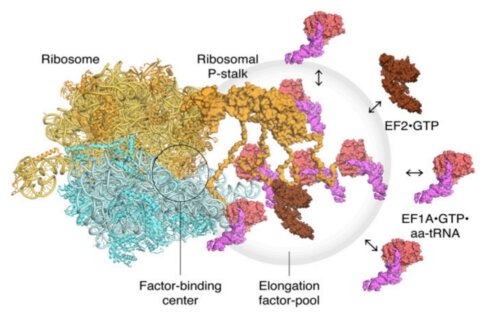
The first step in the mechanism is the recruitment of the factors that are sent to a designated link site located on the main protein, thereby increasing the concentration of the factors at that location, in a mechanism called "pooling of factors" (accumulation of many factors at one point). The second step is binding and stabilizing those functional proteins (translational GTPase) on the binding site in the ribosome in order to accelerate the hydrolysis process of these proteins. From their innovative studies, the scientists were able to obtain visual evidence of the mechanism of protein assembly in the ribosome.
Although the study was unable to provide conclusive evidence as to the activity of the factors once they bind to the ribosome, the researchers did notice that the factors remained in the active area after hydrolysis, a finding that suggests the possible role of the main protein in the subsequent stages of protein synthesis.
The researchers conclude and say: "Future research using high-speed atomic force microscopy (HS-AFM) will provide additional important information for understanding the dynamic activity of these complex translation machines."
A model of the translating ribosomes and the elongation factors. The localized accumulation of translation factors contributes to efficient protein synthesis within a dense intracellular environment.
