A combination of artificial intelligence and protein design algorithms allows for the first time access to a gold mine: millions of natural proteins that can be used in a variety of environmental, medical and industrial applications
Dr. Sheeran Barber-Zucker arrived at the laboratory of Prof. Sheral Fleishman With a dream: she wanted to find a green way to deal with the plastic waste that threatens to drown the world. The initial thought was to draw inspiration from nature - for example, from fungi that cover dead trees and secrete enzymes that cause even the toughest trunks to rot and release nutrients into the soil. Why wouldn't these enzymes roll up their sleeves to help with other crafts like breaking down hard plastics?
However, like dreams, these enzymes are also extremely elusive. "The enzymes called versatile peroxidases are real prima donnas. It is very difficult to work with them", says Prof. Fleischman from the Department of Biomolecular Sciences of the Weizmann Institute of Science. Although in recent years computational methods have been developed in his laboratory that make it possible to make proteins, including enzymes, more stable, or have other desirable properties, but these methods - already used by thousands of research groups around the world - can only be applied when the exact molecular structure of the protein is known. But if the protein is not stable enough to be produced in the laboratory, as in the case of versatile peroxidases, it is easy and impossible to produce crystals from it and bombard them with X-rays - a process known as crystallography and required for deciphering the three-dimensional structure of proteins.
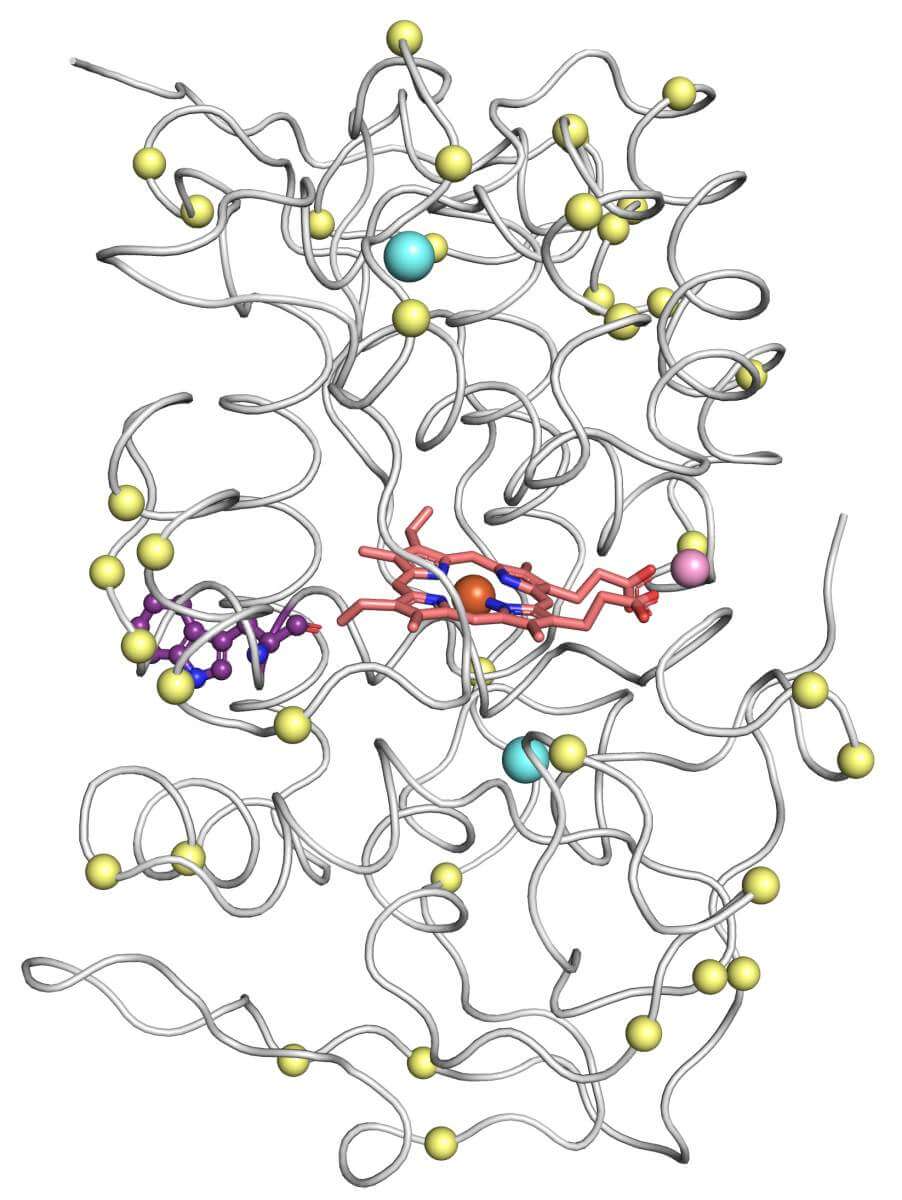
But dreams are not given up so quickly - and Dr. Barber-Zucker insisted on the elusive enzymes. In miraculous timing, the insistence proved itself: since the 80s there have been attempts to skip the need for crystallography and to predict the structure based on the sequence of the amino acids that make up the protein. Until recently, these attempts did not produce particularly reliable results when it comes to complex proteins of the type of versatile peroxidases. However, at the end of 2020, there was a twist in the plot: a few weeks after the start of the project, Dr. Barber-Zucker discovered that a subsidiary of Google, DeepMind, and several university research groups, have greatly improved the prediction capabilities based on artificial intelligence, and that today it is possible to reach prediction results almost as accurate as those reached through crystallography.
The "dark matter" of biology
This technological breakthrough allowed Dr. Barber-Zucker and her colleagues - Vladimir Mindel and Jonathan Yaakov Weinstein, research students in Prof. Fleishman's laboratory, and Prof. Miguel Alcalde and Dr. Eva Garcia Ruiz from the Catalysis Institute in Madrid - achieve results at a speed that was unimaginable before. In fact, until the present study, the structure of only one enzyme from the family of versatile peroxidases has been deciphered - and that too as part of a project that lasted more than a decade and required a team of experts. Now, in less than six months, and without any prior experience or knowledge of this type of enzyme, Dr. Barber-Zucker and her colleagues were able to design, produce and test stable versions of three versatile peroxidases, whose original versions cannot be produced in the laboratory at all.
The combination of AI-based prediction and protein optimization algorithms opens up a huge range of possibilities. "Of the millions of proteins in nature, the structure of less than 0.1% is deciphered. Now we can also study the remaining 99.9% of proteins and use them in medicine and industry," says Prof. Fleishman. "In fact, about half of the proteins in nature cannot be produced in a laboratory at all. These proteins are the 'dark matter' of biology, scientists have so far had no way of determining exactly what they do. In our previous studies, when we were required to design a new protein, we depended on whether the structure of the protein was already deciphered. This question is no longer relevant, we can manage even without the structure. This is a watershed in protein design."
Drug development is one of the fields that can benefit immediately from this development. For example, today, in order to use antibodies derived from laboratory animals to treat humans, it is necessary to invest many resources in the crystallography of the antibodies and the introduction of the necessary structural adjustments. It will now be possible to engineer antibodies much more efficiently.
Environmental applications - the original goal of the scientists - may also benefit from the fruits of the research. For example, the improved enzymes they created may be used to break down agricultural waste and turn it into biofuel - instead of burning or breaking it down with polluting chemicals. In addition, Dr. Barber-Zucker showed that the enzymes she created can break down a certain dye that could become a pollutant, and that it is possible to develop enzymes that can treat different types of environmental pollutants. Moreover, she showed that each of the three enzymes exhibited different activity in the laboratory, that is, it "specialized" in breaking down different components of the wood, and that all three were very stable and resistant to heat - essential qualities for the industry. Based on these findings, Dr. Barber-Zucker aims to develop a "cocktail" of about a dozen enzymes, including the three she created, that will work synergistically to break down wood waste and turn it into biofuels or other useful substances.
And what about the dream of recycling hard plastic waste using enzymes? "It is still a dream, but it may become a reality in the near future," she says.
Another shame on the science website:
