One of the largest proteins of the virus, which the researchers were able to decipher using a new and unique method, is a protein called Nsp2, known as "reluctant" to structural decipherment. Dr. Dina Schneidman: "We now have technology and an experimental protocol to study not only the virus proteins themselves, but also their partners inside the human cell." Prof. Michal Liniel: "Further on, it will be necessary to crack the entire virus in the cellular context in order to simulate all the virus proteins and the interactions between them"
Last June said The former Director General of the Ministry of Health, Moshe Bar Siman Tov, at his graduation ceremony said that "we have learned a lot about the corona virus, but we still don't know anything." Human nature has a hard time coming to terms with the inability to understand the virus." From then until today, about half a year after those things, scientists, researchers and doctors still do not have the full picture of how the internal mechanism of the corona virus works and how it affects the healthy cells, from the inside - that is, what is the role of each protein in the virus, how the protein structures that are formed are organized and how They communicate with each other, which proteins take care of the rapid reproduction of the virus in the cells of the human body, and of course how many mutations of the virus develop in such a short time.
Researchers now know that the COVID-19 corona virus produces in the attacked cell (at least) 29 proteins that are used by it to "hijack" the cellular mechanisms for the purposes of its replication. As in other viruses, the proteins connect to each other and form complex complexes - structures. There are proteins in a virus that have a complete structure, some with a partial structure and some without a structure. In many laboratories, during the past year, they tried to decipher and understand the atomic structure of each of these proteins separately, in order to understand the complete activity of the virus. Understanding the atomic structure is necessary in most cases for the design of drugs and other strategies to fight the virus.
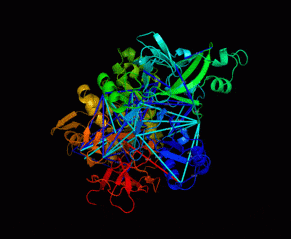
Nsp2 – the recalcitrant protein
One of the largest proteins of the virus, a protein called Nsp2, was found to be "recalcitrant" to structural decipherment. This was particularly surprising because it is a protein that on the surface has "normal" characteristics (a soluble, non-membrane protein, with a continuous structure), similar to thousands of other proteins regularly deciphered by crystallography or freeze electron microscopy. In addition, almost nothing is known about its role in the reproduction of the virus. The protein aroused the curiosity of a number of researchers from the Hebrew University, among them the researcher of corona viruses from the Institute of Life Sciences Prof. Michal Lineal, Dr. Dina Schneidman from the Department of Biological Chemistry and the School of Computer Sciences, and even Dr. Nir Klisman from the Institute of Life Sciences.
Following this, it was decided to form an extensive team (12 female researchers, 2 male researchers) from the Hebrew University of Jerusalem and Hadassah College, which will try to lead to the solution of the mystery surrounding the Nsp2 protein, and other proteins in the virus. The team was divided into research subgroups that performed various actions - from growing cells that produce the proteins, performing mass spectroscopy to calculating structural models for the corona proteins. The new study carried out by the researchers Published at this time on bioRxiv.org, used to publish scientific articles before they have been peer-reviewed. It is primarily designed to understand what the virus does inside the infected cell through the investigation of the structure of several virus proteins in human cells, including Nsp2, Nsp1 and Nucleocapsid protein (known as N for short).
The researchers hypothesized that Nsp2 is unstable outside the cell in which it is expressed, and in such a case, the usual methods for determining its structure will fail, because they are all based on taking the protein out of the cell and purifying it by biochemical means. They circumvented this limitation by using an intracellular method to study the structure. A chemical compound was introduced into the living cell which irreversibly binds two sites in the protein to each other (chemical cross-linking), only if they are physically close in the three-dimensional structure. Only after the chemical action was finished, the researchers broke the cell and isolated the Nsp2 protein from it. With the help of a device known as a mass spectrometer, it was possible to identify which regions of the protein were close to each other and passed the chemical cross-linking step. More than 40 such points of proximity were identified in the studied protein who testified that The long protein chain of the Nsp2 has a particularly convoluted and complex structure.
Google's DEEP MIND helped decipher
Because of the long sequence and the complex structure of Nsp2, the forty "clues" about its structure were not enough to fully decipher and understand the atomic structure, so the researchers had to use additional sources. In perfect timing, this year a subsidiary of Google DeepMind developed a revolutionary and successful software for predicting the structure of proteins, called AlphaFold2. One of the challenging proteins, whose structure the software recently tried to find, was fortunately Nsp2. The software was unable to predict the full structure, but uploaded to the network several substructures for narrower regions of the Nsp2 sequence. This was enough to fill in the missing information for the researchers. Combining the substructures from AlphaFold2 with proximity clues from the chemical cross-linking method made it possible to produce the complete atomic structure.
Investigation of the new structure showed that it contains three zinc binding sites. Zinc is an essential ion for the operation of the replication machine of the genetic code of the virus. It was known that inside the attacked cell the virus creates a kind of isolated complexes in which the replication machines work separately from the rest of the cell. Also, previous work has shown that Nsp2 is transported to these isolated replication regions. Therefore, the researchers hypothesized that one of the functions of Nsp2 is to ensure the supply of zinc in the isolated replication regions. "Apparently the protein supplies zinc to the unique organelle in the cell cytoplasm, where the virus replicates inside the cell," the researchers clarified.
The structure of the studied protein provides another and interesting point of view on the accelerated evolutionary development of the current virus. In the corona viruses known to man, such as those that cause the common cold every winter, there is only one zinc binding site. In contrast, in the current subfamily of viruses (which caused the SARS epidemic in 2002 in Asia, and are responsible for the current epidemic), three binding sites were discovered in the study, and there are structural hints that a fourth binding site is developing and shaping in the SARS-COV-2 virus.
The researchers used the same method to study two other proteins in the corona virus - the protein packaging the genetic material (Nucleocapsid protein or simply N), and a protein called Nsp1 which is one of the main proteins of the virus in evading the immune system following infection. Dr. Klisman, who took a central part in the research, explains that "even in these two cases, we discovered that types of structures are formed inside the cell that until now have not been able to be thoroughly investigated with extracellular methods. The cell creates a very complex environment, which is very difficult for us to reproduce in vitro"; Dr. Schneidman concludes: "The method we used looks very promising and is suitable for examining many proteins. While the virus can manage without Nsp2, Nsp1 is an essential protein in the corona virus and with its help it manages to trick the immune system. Now we have the technology and experimental protocol to continue to study not only the virus proteins themselves, but mainly their partners inside the human cell. The road is still long and very winding, but it started with good news"; And Prof. Liniel clarifies that "this is just the beginning. In the future, it will be necessary to study the entire virus in the cellular context in order to simulate all the virus proteins and the interactions between them."
