While this assessment of an evolutionary relationship between vertebrates and echinoderms has become consensus, unexpected findings from comparative living DNA studies designed to reconstruct evolutionary trees are calling the consensus into question.
By: Max Telford, Professor of Zoology, UCL and Faslia Capley, Research Fellow in Genetics, Evolution and Environment, UCL. Translated by Avi Blizovsky
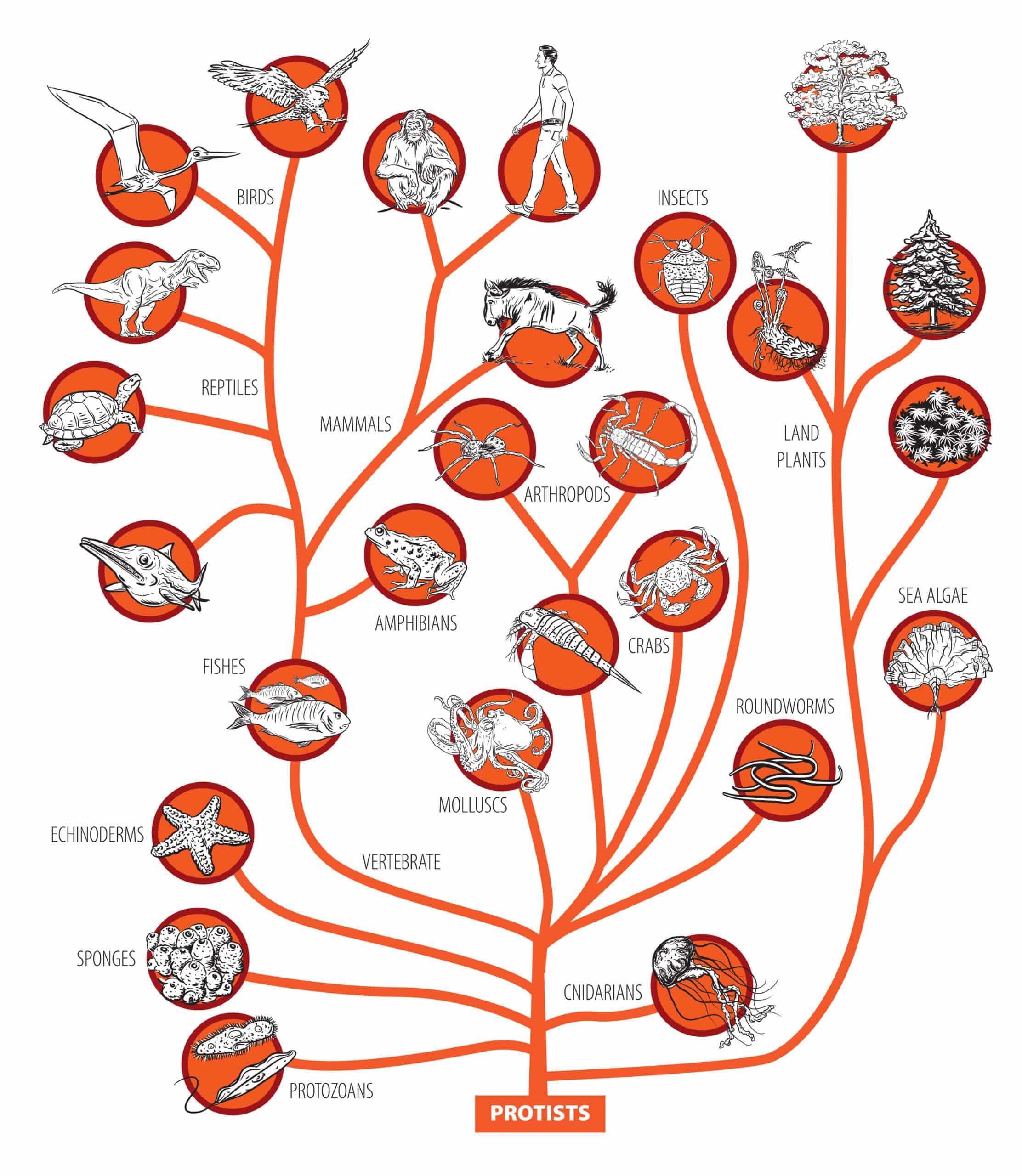
The question of how humans evolved from the first animals during the last 600 million years is a fascinating question. When piecing together the many steps leading from the first simple life form that formed our ancestor to modern Homo sapiens, the first thing we need to know is how are we related to other animal groups?
Many aspects of our family tree are self-evident. It is easy to see that we belong to the group of great apes, primates and mammals, for example. Delving into our history, even Aristotle was aware that mammals, birds, reptiles, amphibians and fish are united by the common feature of a backbone in the group we call vertebrates.
The origins of vertebrates lie deep in the history of animal life on Earth, and the earliest dated fossils showing any backbone are over half a billion years old. There are inherent difficulties in dissolving such ancient relationships. Scientists have struggled with many aspects of them for the past 150 years. Now our new study, published in Science Advances, offers insight.
Relatives of the starfish?
Given these difficulties, the fact that there has been a long period of consensus about the identity of vertebrate relatives is highly unusual. The general perception for over a century has been that, surprisingly, the closest animals among the non-vertebrates are a group that bears no striking resemblance to vertebrates at all. they the skin strands - a group that includes starfish, sea cucumbers and sea urchins.
In 1908 an Austrian zoologist named Karl Gruben proposed this connection. Groven examined the earliest events of embryonic development in different groups of species (how the first cells of the embryo divide and how the mouth first forms) with the aim of splitting the main part of the animal kingdom into two large branches. He claimed that vertebrates and echinoderms are part of a group called "deuterostomes" and that the other invertebrates in the group known as protostomes are insects, earthworms, molluscs and nematodes.
While this assessment of an evolutionary relationship between vertebrates and echinoderms has become consensus, unexpected findings from comparative living DNA studies designed to reconstruct evolutionary trees are calling the consensus into question. At the same time, some of the specific features of the way embryos develop that were highlighted as special to the deuterostome branch of animals were discovered in several species of protostomes.
These findings suggest that the evidence that echinoderms are the closest vertebrate relatives may be weaker than long believed.
New findings
We set out to check the evidence. Changes in the DNA sequences of the genes occurred in different lineages of the tree of life - and a record of the relationships between them. We used a computer to compare the DNA sequences of genes from all over the animal kingdom to reconstruct the animal evolutionary tree.
To see if the DNA data convincingly supports a close relationship between vertebrates and invertebrates, we looked at the number of DNA changes also found in vertebrates and skin strands but not in other animals. Species with these common traits, if found, would be evidence supporting the close relationship between the species.
Drawings of the three possible trees. Max Telford, courtesy of the author
If you look at about 5,000 different genes, about 70% of the animals in the protostome branch had more unique changes than the deuterostome branch. That is, the animals on the protostome branch share a lot of unique changes in their DNA - so this branch is well supported by the DNA evidence. The close relationship between the vertebrates, however, is supported by much weaker evidence - they share relatively few unique changes in DNA.
After that we tried different arrangements of the vertebrates, skin strands and protostomes: first, the classic tree linking vertebrates and skin strands; secondly, the tree where the strands of skin are actually more related to protostomes than to vertebrates; and finally a tree in which the vertebrates are more related to protostomes than to skin spines
We saw that many genes did not have a huge amount of DNA variation supporting any of the three trees. Of the gene analyzes that showed evidence that clearly favored one of the three alternatives, a very small majority of genes showed stronger evidence for the deuterostome tree than for the other two options. Our result indicates that the three branches - vertebrates, echinoderms and protostomes - separated from each other in a short period of time, meaning that there was not much time to accumulate many changes in DNA. This means that it is now very difficult to understand which of the three possible trees referring to these groups are correct.
The deuterostome tree
So why did most previous DNA studies support a tree with the deuterostome group, despite our experiments showing that there is almost nothing that distinguishes this tree from the two alternatives? We wanted to find out if the analyzes are prone to errors that can arise when different branches developed at different speeds.
To find out, we used a computer to simulate the evolution of DNA according to each scenario. We started with a random synthetic DNA sequence representing an ancient animal. This DNA scrub was made possible by the accumulation of mutations according to each of the three trees.
When we ran data against the deuterostome tree, we always sorted the tree. However, when we ran data that fit the other two trees, we didn't always copy the correct tree - sometimes we created the deuterostome tree by mistake (add 'on purpose', if I understand their intent correctly). This mistake was especially common when we changed the rates of evolution in the different groups.
This result suggests that there could be an error in the actual DNA data that would cause us to reconstruct the deuterostome tree, even if it was incorrect. The error may be caused by the two deuterostome groups (vertebrates and echinoderms) being tempted at a slower rate than the other groups. This would make them look more similar to each other than to protostomes, even if they are not really related to each other.
Our conclusion is that the confidence in close relationships between vertebrates and invertebrates - in textbooks for more than a century - is out of place. We have shown that this evolutionary problem is particularly difficult to solve and that we vertebrates may turn out to be more closely related to snails and flies than to starfish.
For an article in The Conversation
More of the topic in Hayadan:
- The tree of life containing the connections between the 2.3 million species known to science - from bacteria to humans - has been mapped
- Tree of Life
- A billion-year arms race against viruses has shaped our evolution
- The genome of a common octopus species, which allows them to have three almost completely independent hearts and minds in each arm, has been deciphered

2 תגובות
What is interesting and perhaps strange is that apart from fish (not fishes) there is not a single species on a branch, meaning that all the ancestors of the species from which the species split were extinct or unknown to science.
It seems that the problem is very difficult to solve, especially since there is lateral transfer of genes between different species - the CRISPR system is known in bacteria that may have been or still have remnants of it in multicellular organisms, there is transfer by viruses, and much more is unknown. So to tap evolution for hundreds of millions of years from today is a bit of an attempt to hit a target too far.