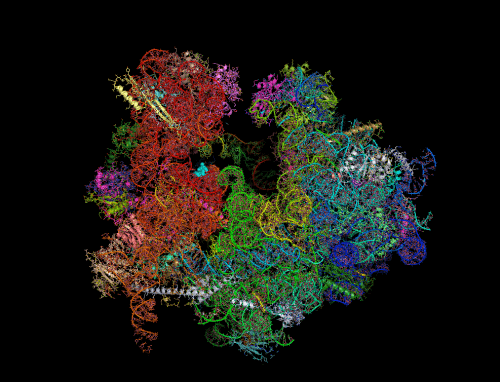
Ribosomes - the cell's protein production machines - are a central battleground in the fight against harmful bacteria. Many antibiotics disable the activity of bacterial ribosomes by binding to their active sites. Many times bacteria develop resistance to antibiotics through changes that prevent the drug from binding to the ribosome. A new study by Weizmann Institute scientists, SRecently published in the scientific journal Nature Communications., may point to a lesser-known way to make bacteria more sensitive to antibiotics - by preventing the natural deactivation mechanism of the ribosomes.
In bacterial cells, especially in stress situations, pairs of ribosomes are sometimes formed that have joined each other through certain sites on the smaller of the two subunits of the ribosome. When two ribosomes stick together in this way, their activity is disabled and they enter a kind of "dormancy". This is a temporary condition that can occur, for example, following a lack of nutrients or sometimes even as part of the cell's normal activity mechanism. The pairs of ribosomes can "wake up" from this slumber within seconds and resume protein production.
How do two ribosomes join? This process has been documented in the common gram-negative laboratory bacterium E. coli, but gram-positive bacteria may use other mechanisms. Among these bacteria is also Staphylococcus aureus, which can cause life-threatening diseases. Understanding the dormancy process in these bacteria, including resistant strains of the bacteria, may provide crucial insights for the development of future drugs.
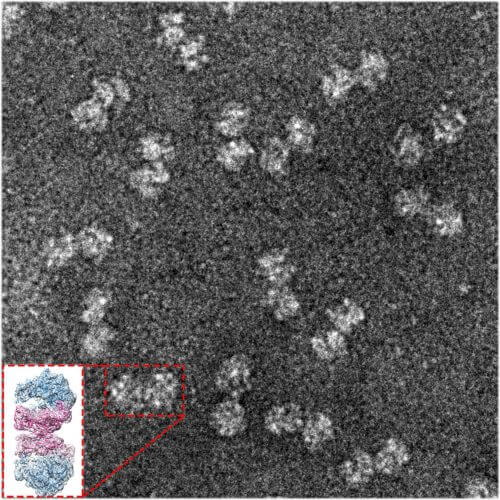
To answer the question, researchers from St. Louis University, Missouri, turned to the research group of Prof. Ada Yonathin the Department of Structural Biology at the Weizmann Institute of Science due to her unique expertise in revealing the exact XNUMXD structure of the ribosome which contributes to understanding its function. The research was led by research student Donna Matzov from Prof. Yonat's laboratory, who, using sensitive purification methods, separated the ribosomes from the cells, and later separated the dormant ribosomes from the other unpaired ribosomes.
"The method we used is electron microscopy of single particles in deep freezing, for which the Nobel Prize in Chemistry was awarded this year. It is this method that allowed us to discern the exact details of dormant ribosomes"
Subsequently, a microscopic analysis was conducted with high resolution and XNUMXD detail, in collaboration with the electron microscopy group of Dr. Alexey Amonts from the University of Stockholm, Sweden - where the dormant ribosomes are positioned for flight. Advanced computer algorithms that the researchers applied to the imaging products revealed with great precision the role of a non-ribosomal cellular protein, called hibernating promoting factor ('hibernation promoting factor', or HPF), in creating the connection between the two ribosomes. In gram-negative bacteria such as E. coli, a version of this protein resides near the active site on the ribosome. Together with another protein called RMF, HPF activates in Gram-negative bacteria a chain of events that leads both to the external coupling of two ribosomes and to the cessation of protein production within the active site of each of the ribosomes. However, the RMF is absent from Gram-positive bacteria, and their HPF protein is twice as long. "At first we thought that HPF might be a hybrid protein, with an extension corresponding to the missing RMF," Matzov says.
However, in practice, the researchers discovered a completely different mechanism: the HPF of dormant ribosomes in Gram-positive bacteria is a relatively long protein with two active sites, one at each end, connected by a flexible loop. Instead of causing a domino effect, it acts directly: one end reaches into the site of protein production, where it blocks activity, while the other end reaches the outside of the small subunit, where it makes direct contact with the corresponding HPF protein in the ribosome The second. After this coupling occurs, other parts of the conjugated ribosomes can connect to a communication network between the two, thus helping to maintain the paired ribosomal structure and stay informed of changes that could allow them to resume their activity.

Dr. Anat Bashan from Yonat's laboratory explains: "The method we used is electron microscopy of single particles in deep freezing (cryo-EM), for which this year the Nobel Prize in Chemistry was awarded. It was this method that allowed us to discern the exact details of dormant ribosomes. Contrary to traditional crystallographic methods, we were not required to create ribosome crystals in order to determine the structure, and therefore we were able to study many samples - configurations and dynamics."
"Understanding the role of the HPF protein may be the first step in developing a way to prevent the hibernation of ribosomes, thus making them more sensitive to antibiotics," Matzov says. "And since such a method only targets Gram-positive bacteria, many friendly bacteria in our bodies may avoid harm."
Dr. Ella Zimmerman from Yonat's research group also participated in the study; Dr. Shintaro Ibera from Stockholm University; and Dr. Arnav Basu and Prof. Mi-Ngan Yap from the St. Louis University School of Medicine.
